Probe CUST_31135_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31135_PI426222305 | JHI_St_60k_v1 | DMT400063936 | GCAAAGTATGTGTGTTCTAAAAGGCCCGAGTTTGCTTAACATATTGTGTAATAAACTACT |
All Microarray Probes Designed to Gene DMG400024849
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_31213_PI426222305 | JHI_St_60k_v1 | DMT400063937 | GCAAAGTATGTGTGTTCTAAAAGGCCCGAGTTTGCTTAACATATTGTGTAATAAACTACT |
| CUST_31135_PI426222305 | JHI_St_60k_v1 | DMT400063936 | GCAAAGTATGTGTGTTCTAAAAGGCCCGAGTTTGCTTAACATATTGTGTAATAAACTACT |
Microarray Signals from CUST_31135_PI426222305
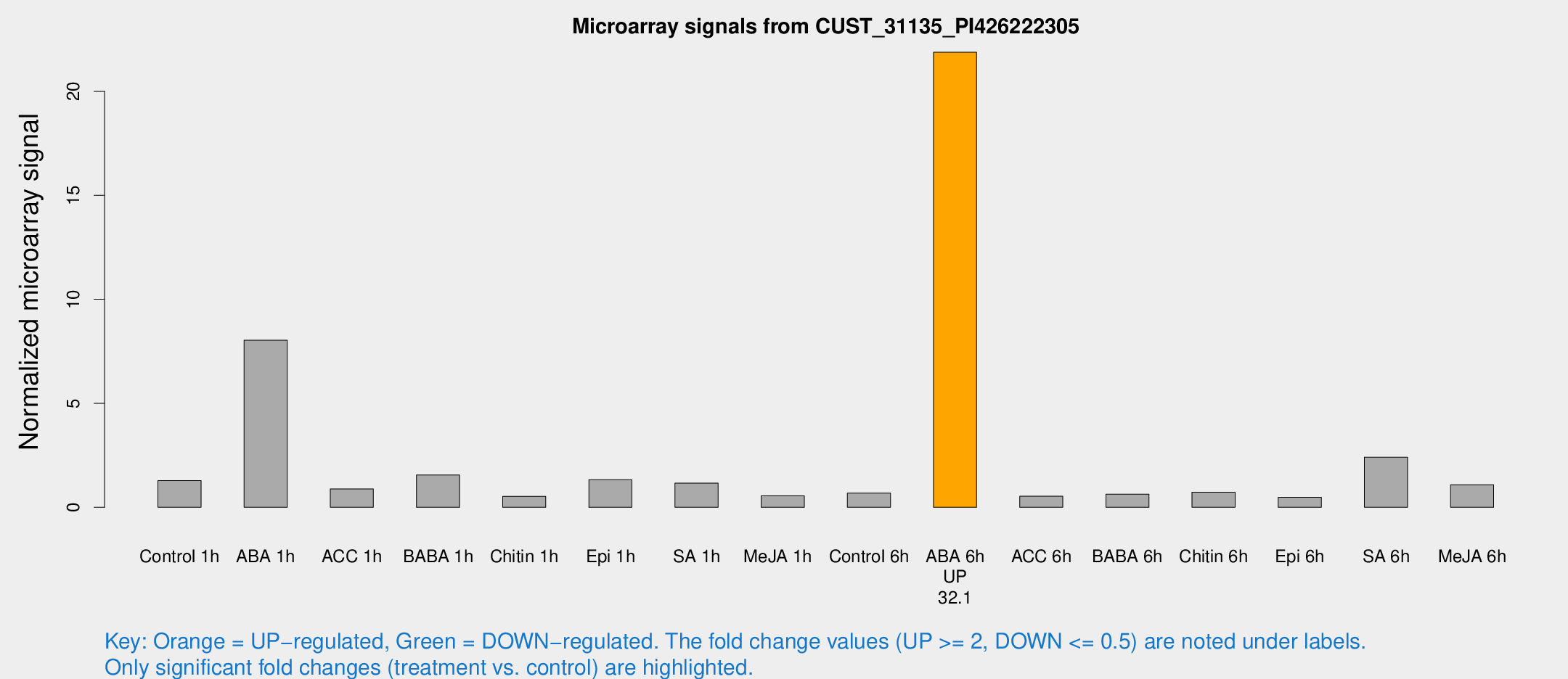
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 38.8117 | 5.56515 | 1.28561 | 0.287355 |
| ABA 1h | 234.403 | 71.5741 | 8.03261 | 2.67803 |
| ACC 1h | 29.1164 | 7.52236 | 0.880403 | 0.215929 |
| BABA 1h | 45.6999 | 8.4707 | 1.55175 | 0.177958 |
| Chitin 1h | 17.7234 | 8.3811 | 0.529717 | 0.269853 |
| Epi 1h | 34.3515 | 3.91665 | 1.3274 | 0.156979 |
| SA 1h | 36.7104 | 7.65658 | 1.1616 | 0.228663 |
| Me-JA 1h | 14.4913 | 3.75359 | 0.550389 | 0.246886 |
| Control 6h | 20.5541 | 3.82037 | 0.681572 | 0.149345 |
| ABA 6h | 724.961 | 152.862 | 21.8799 | 4.3235 |
| ACC 6h | 19.0303 | 4.39994 | 0.533865 | 0.161763 |
| BABA 6h | 21.8605 | 5.20958 | 0.627845 | 0.132507 |
| Chitin 6h | 23.4405 | 4.2436 | 0.724056 | 0.14137 |
| Epi 6h | 20.0528 | 8.49359 | 0.484564 | 0.324823 |
| SA 6h | 122.931 | 87.9597 | 2.41352 | 2.57242 |
| Me-JA 6h | 37.201 | 14.8192 | 1.08778 | 0.35169 |
Source Transcript PGSC0003DMT400063936 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G77120.1 | +1 | 0.0 | 626 | 313/369 (85%) | alcohol dehydrogenase 1 | chr1:28975509-28977216 FORWARD LENGTH=379 |
