Probe CUST_30964_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30964_PI426222305 | JHI_St_60k_v1 | DMT400095861 | ACCTCATCTGGCTTGATCACTCCAGTTTTTATAGAGGATCAATTCATTAATGTATGGTAA |
All Microarray Probes Designed to Gene DMG400045432
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30964_PI426222305 | JHI_St_60k_v1 | DMT400095861 | ACCTCATCTGGCTTGATCACTCCAGTTTTTATAGAGGATCAATTCATTAATGTATGGTAA |
Microarray Signals from CUST_30964_PI426222305
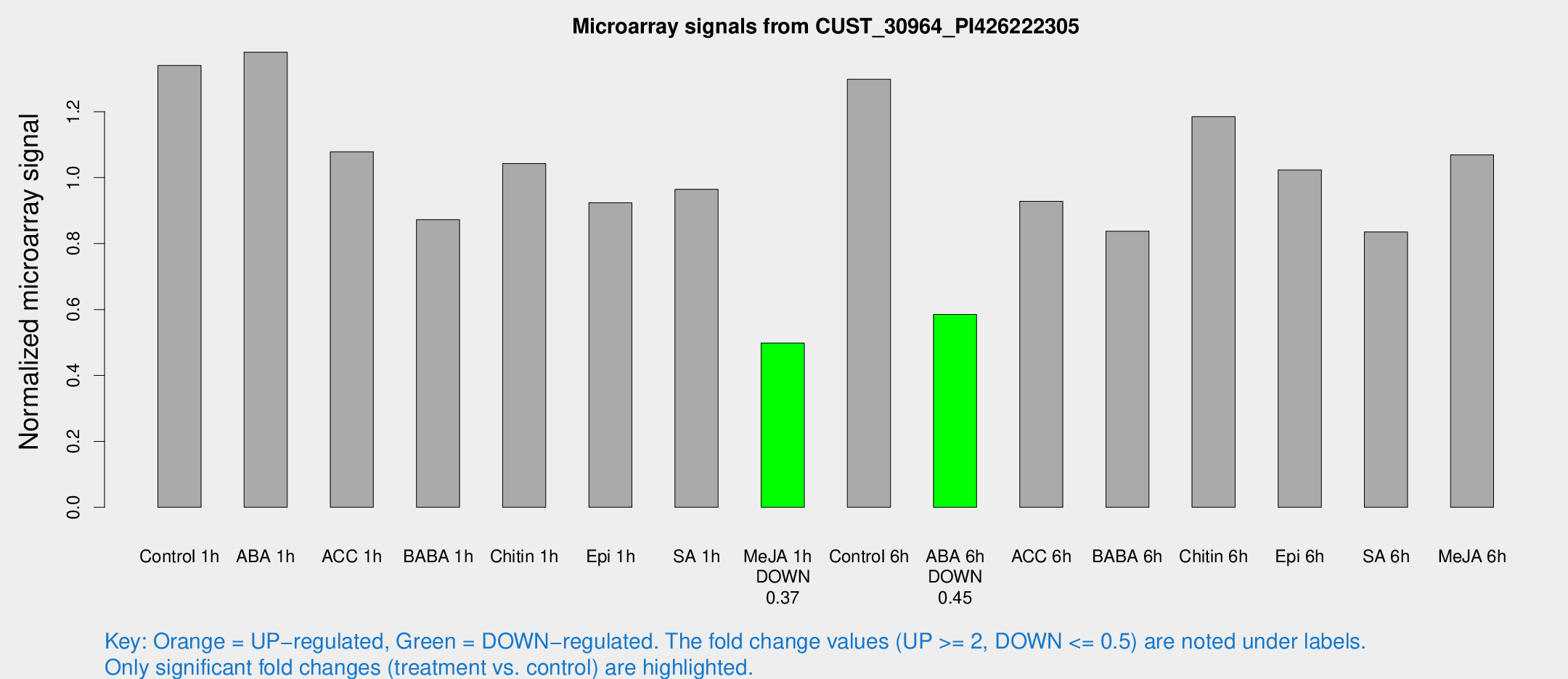
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3266.24 | 362.314 | 1.34031 | 0.0879672 |
| ABA 1h | 2949.09 | 193.152 | 1.38032 | 0.140863 |
| ACC 1h | 2832.57 | 629.063 | 1.07835 | 0.201283 |
| BABA 1h | 2072.35 | 318.272 | 0.872398 | 0.0647835 |
| Chitin 1h | 2263.66 | 142.71 | 1.04264 | 0.0602146 |
| Epi 1h | 1923.23 | 111.094 | 0.924023 | 0.053369 |
| SA 1h | 2463.29 | 419.377 | 0.964715 | 0.141125 |
| Me-JA 1h | 979.078 | 56.6938 | 0.498098 | 0.0288038 |
| Control 6h | 3205.32 | 466.641 | 1.29852 | 0.0825552 |
| ABA 6h | 1500.65 | 91.9404 | 0.584909 | 0.033796 |
| ACC 6h | 2629.86 | 444.668 | 0.928394 | 0.0536185 |
| BABA 6h | 2313.93 | 389.852 | 0.837453 | 0.130861 |
| Chitin 6h | 3039.92 | 175.977 | 1.18485 | 0.094318 |
| Epi 6h | 2815.89 | 324.578 | 1.02357 | 0.200148 |
| SA 6h | 2088.74 | 445.709 | 0.835392 | 0.103371 |
| Me-JA 6h | 2650.23 | 484.955 | 1.06913 | 0.110482 |
Source Transcript PGSC0003DMT400095861 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G10560.1 | +1 | 4e-139 | 423 | 235/553 (42%) | plant U-box 18 | chr1:3484613-3486706 FORWARD LENGTH=697 |
