Probe CUST_30912_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30912_PI426222305 | JHI_St_60k_v1 | DMT400041410 | TACTGGTGTCTTGACAGAACAACAACGGCTCATATGCCGTGGAAAAGTGTTGAAGGATGA |
All Microarray Probes Designed to Gene DMG401016046
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30912_PI426222305 | JHI_St_60k_v1 | DMT400041410 | TACTGGTGTCTTGACAGAACAACAACGGCTCATATGCCGTGGAAAAGTGTTGAAGGATGA |
Microarray Signals from CUST_30912_PI426222305
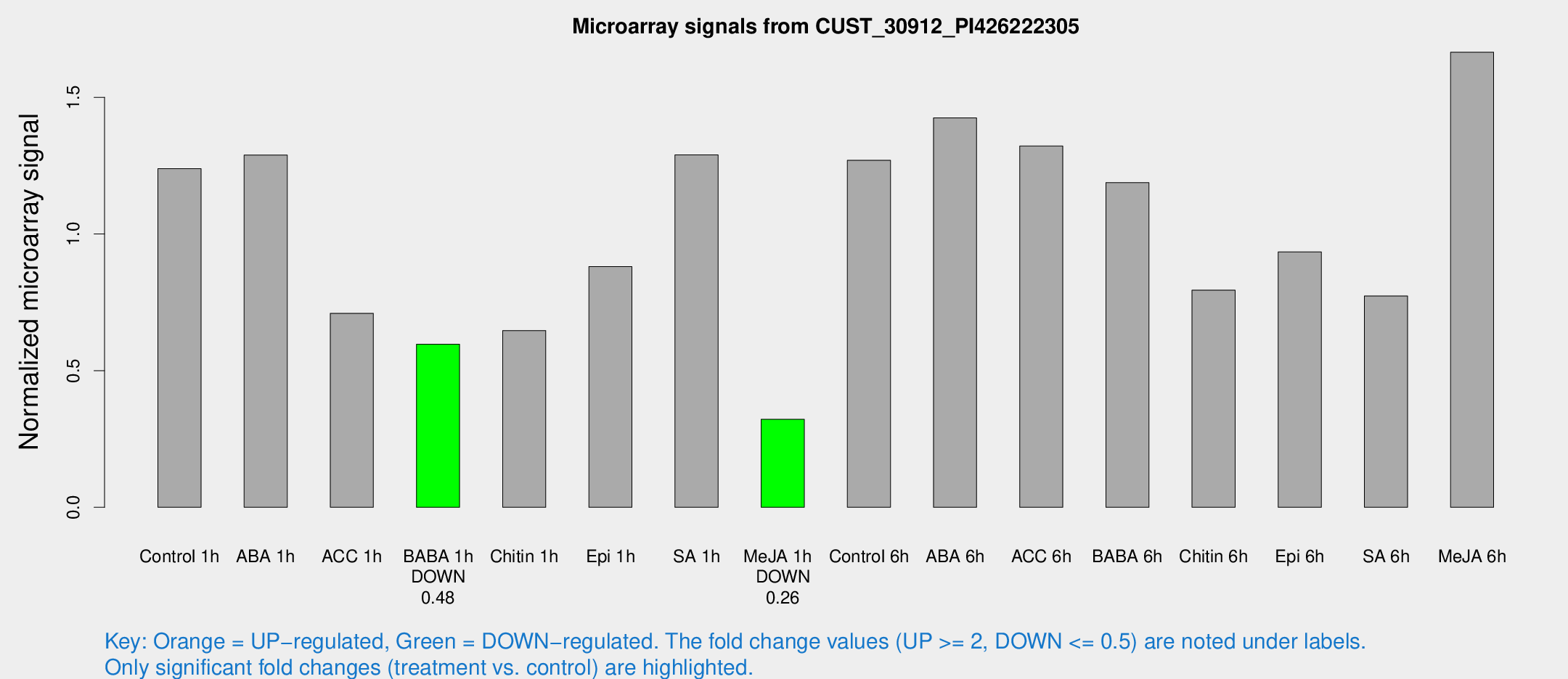
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 38.8461 | 3.85385 | 1.23875 | 0.123025 |
| ABA 1h | 36.0811 | 4.19447 | 1.28897 | 0.204344 |
| ACC 1h | 25.0586 | 6.66954 | 0.709809 | 0.188299 |
| BABA 1h | 18.4277 | 3.48373 | 0.596426 | 0.119238 |
| Chitin 1h | 18.4201 | 3.37606 | 0.646748 | 0.12305 |
| Epi 1h | 24.28 | 3.41735 | 0.880801 | 0.132087 |
| SA 1h | 44.3452 | 11.1365 | 1.28954 | 0.27667 |
| Me-JA 1h | 8.64434 | 3.28516 | 0.321883 | 0.133932 |
| Control 6h | 53.7082 | 22.2896 | 1.26968 | 0.852934 |
| ABA 6h | 51.8036 | 13.8108 | 1.42465 | 0.426423 |
| ACC 6h | 49.7271 | 10.772 | 1.32177 | 0.14894 |
| BABA 6h | 54.5175 | 21.0928 | 1.18791 | 0.851259 |
| Chitin 6h | 36.9311 | 15.0915 | 0.794485 | 0.78457 |
| Epi 6h | 36.0811 | 11.4938 | 0.934655 | 0.285653 |
| SA 6h | 29.7948 | 12.0692 | 0.772998 | 0.539357 |
| Me-JA 6h | 62.4992 | 21.5405 | 1.66502 | 0.757246 |
Source Transcript PGSC0003DMT400041410 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G25270.1 | +1 | 2e-37 | 143 | 74/109 (68%) | Ubiquitin-like superfamily protein | chr5:8757577-8762002 REVERSE LENGTH=658 |
