Probe CUST_30819_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30819_PI426222305 | JHI_St_60k_v1 | DMT400046207 | TTTCTCCAACTGCAAAAATATCTCTGCTTTTGTTGTAGACTCTGTTTCTGTAGCTATTCA |
All Microarray Probes Designed to Gene DMG400017936
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30819_PI426222305 | JHI_St_60k_v1 | DMT400046207 | TTTCTCCAACTGCAAAAATATCTCTGCTTTTGTTGTAGACTCTGTTTCTGTAGCTATTCA |
Microarray Signals from CUST_30819_PI426222305
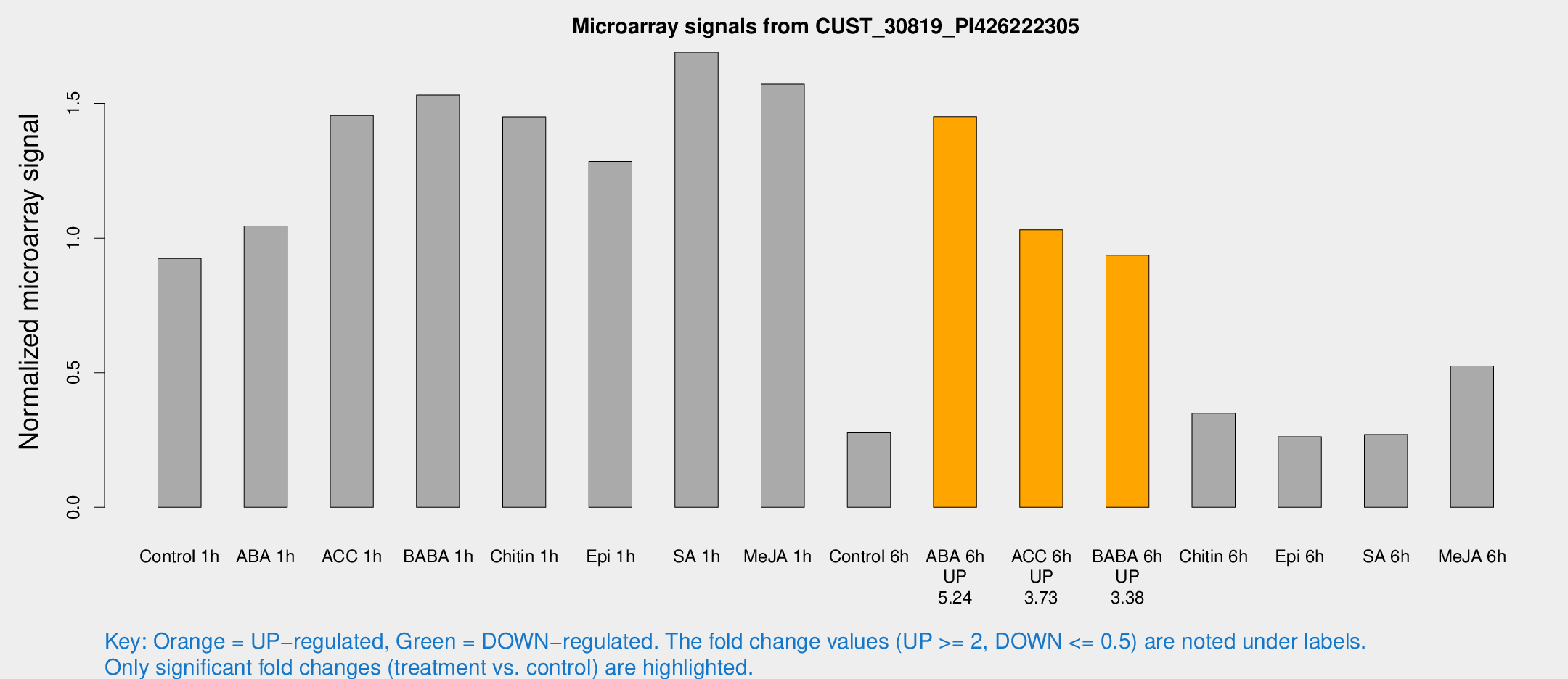
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6703.34 | 388.327 | 0.924139 | 0.0566615 |
| ABA 1h | 7169.61 | 1695.02 | 1.04493 | 0.251061 |
| ACC 1h | 12696.2 | 4203.52 | 1.45503 | 0.583663 |
| BABA 1h | 11479.8 | 3001.73 | 1.53115 | 0.290368 |
| Chitin 1h | 9490.39 | 766.932 | 1.45045 | 0.110857 |
| Epi 1h | 8129.09 | 866.863 | 1.28505 | 0.156125 |
| SA 1h | 13044 | 2353.47 | 1.69024 | 0.255184 |
| Me-JA 1h | 9400.03 | 1090.13 | 1.57168 | 0.0907427 |
| Control 6h | 2235.39 | 636.99 | 0.276758 | 0.0744111 |
| ABA 6h | 12039.3 | 2926.15 | 1.451 | 0.381603 |
| ACC 6h | 8820.67 | 1444.22 | 1.0311 | 0.183848 |
| BABA 6h | 8325.67 | 2639.6 | 0.936402 | 0.278646 |
| Chitin 6h | 2711.33 | 272.263 | 0.348886 | 0.0425881 |
| Epi 6h | 2158.08 | 218.19 | 0.262179 | 0.027737 |
| SA 6h | 1955.3 | 206.857 | 0.270499 | 0.0239238 |
| Me-JA 6h | 3893.59 | 670.976 | 0.525106 | 0.0467946 |
Source Transcript PGSC0003DMT400046207 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G02380.1 | +1 | 4e-14 | 67 | 48/96 (50%) | senescence-associated gene 21 | chr4:1046414-1046807 REVERSE LENGTH=97 |
