Probe CUST_30793_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30793_PI426222305 | JHI_St_60k_v1 | DMT400037956 | TTGCGTGTAATTATTTCTTGTTCTTCTAAGAGAATGTATCTCAACTTTCCATTGGTGGTG |
All Microarray Probes Designed to Gene DMG400014641
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30793_PI426222305 | JHI_St_60k_v1 | DMT400037956 | TTGCGTGTAATTATTTCTTGTTCTTCTAAGAGAATGTATCTCAACTTTCCATTGGTGGTG |
Microarray Signals from CUST_30793_PI426222305
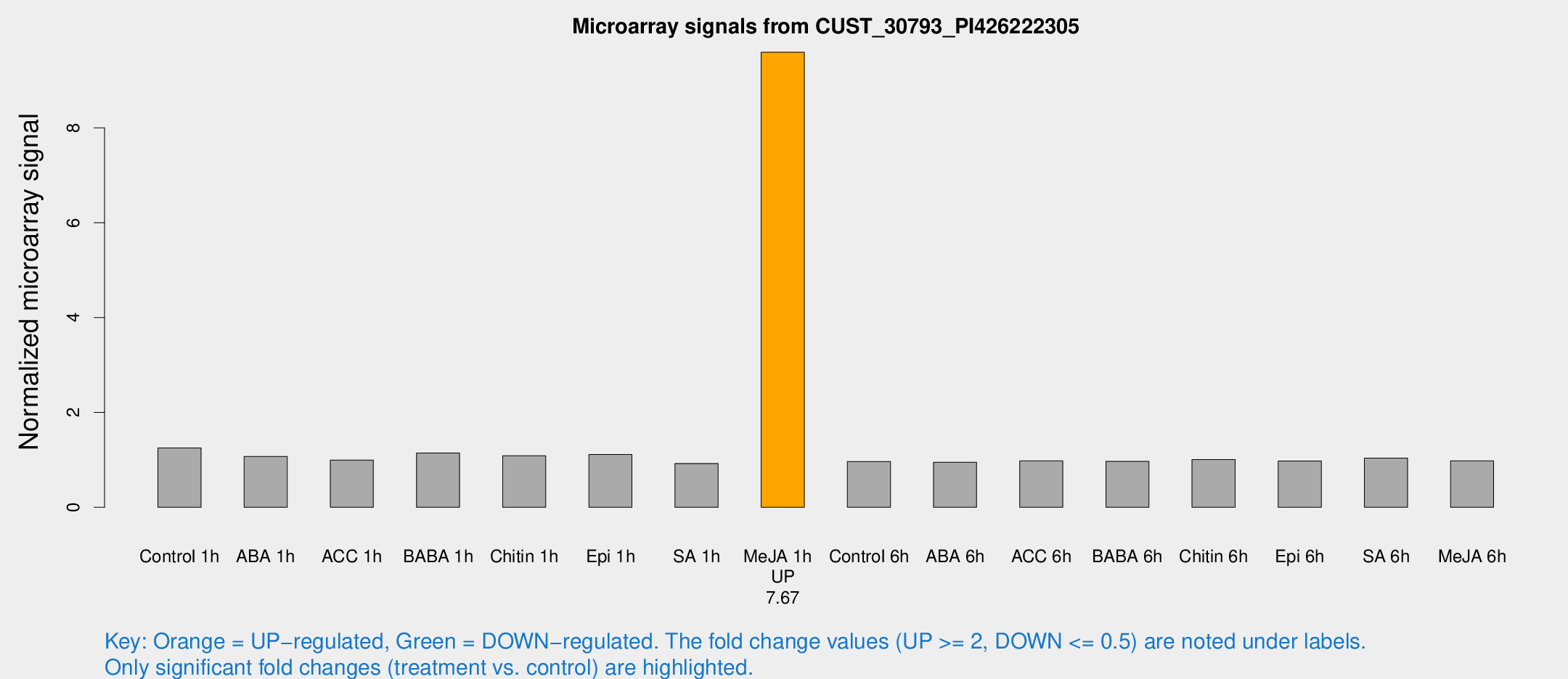
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.02324 | 2.88938 | 1.25082 | 0.588635 |
| ABA 1h | 4.74292 | 2.74855 | 1.07269 | 0.58927 |
| ACC 1h | 5.40633 | 3.13876 | 0.997086 | 0.577731 |
| BABA 1h | 5.91169 | 3.03746 | 1.14408 | 0.603585 |
| Chitin 1h | 4.93839 | 2.8717 | 1.0869 | 0.602275 |
| Epi 1h | 4.94136 | 2.87303 | 1.11586 | 0.628454 |
| SA 1h | 4.969 | 2.87749 | 0.923701 | 0.531585 |
| Me-JA 1h | 41.8286 | 4.96074 | 9.59531 | 1.93908 |
| Control 6h | 5.09133 | 2.94888 | 0.966381 | 0.558175 |
| ABA 6h | 5.31297 | 3.08188 | 0.950397 | 0.548861 |
| ACC 6h | 6.05063 | 3.58621 | 0.97909 | 0.566962 |
| BABA 6h | 5.72063 | 3.32214 | 0.968071 | 0.561051 |
| Chitin 6h | 5.64888 | 3.27161 | 1.00684 | 0.582985 |
| Epi 6h | 5.84887 | 3.416 | 0.976853 | 0.56616 |
| SA 6h | 5.40191 | 3.12988 | 1.03582 | 0.599823 |
| Me-JA 6h | 5.07994 | 2.94753 | 0.979004 | 0.560988 |
Source Transcript PGSC0003DMT400037956 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G19260.1 | +2 | 4e-20 | 88 | 62/236 (26%) | Protein of unknown function (DUF3049) | chr5:6479494-6480360 REVERSE LENGTH=288 |
