Probe CUST_30338_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30338_PI426222305 | JHI_St_60k_v1 | DMT400069946 | CGAGCAATTGGTAGGCATATACTTCTACTGGATCTAAAATTGTGAAGCAAAAATGAAATG |
All Microarray Probes Designed to Gene DMG400027196
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30338_PI426222305 | JHI_St_60k_v1 | DMT400069946 | CGAGCAATTGGTAGGCATATACTTCTACTGGATCTAAAATTGTGAAGCAAAAATGAAATG |
| CUST_30314_PI426222305 | JHI_St_60k_v1 | DMT400069947 | TGGCGACAATTATTTGAAACCATATGTTATATCAGAACCGGAGGTGACGATAACAGATCG |
Microarray Signals from CUST_30338_PI426222305
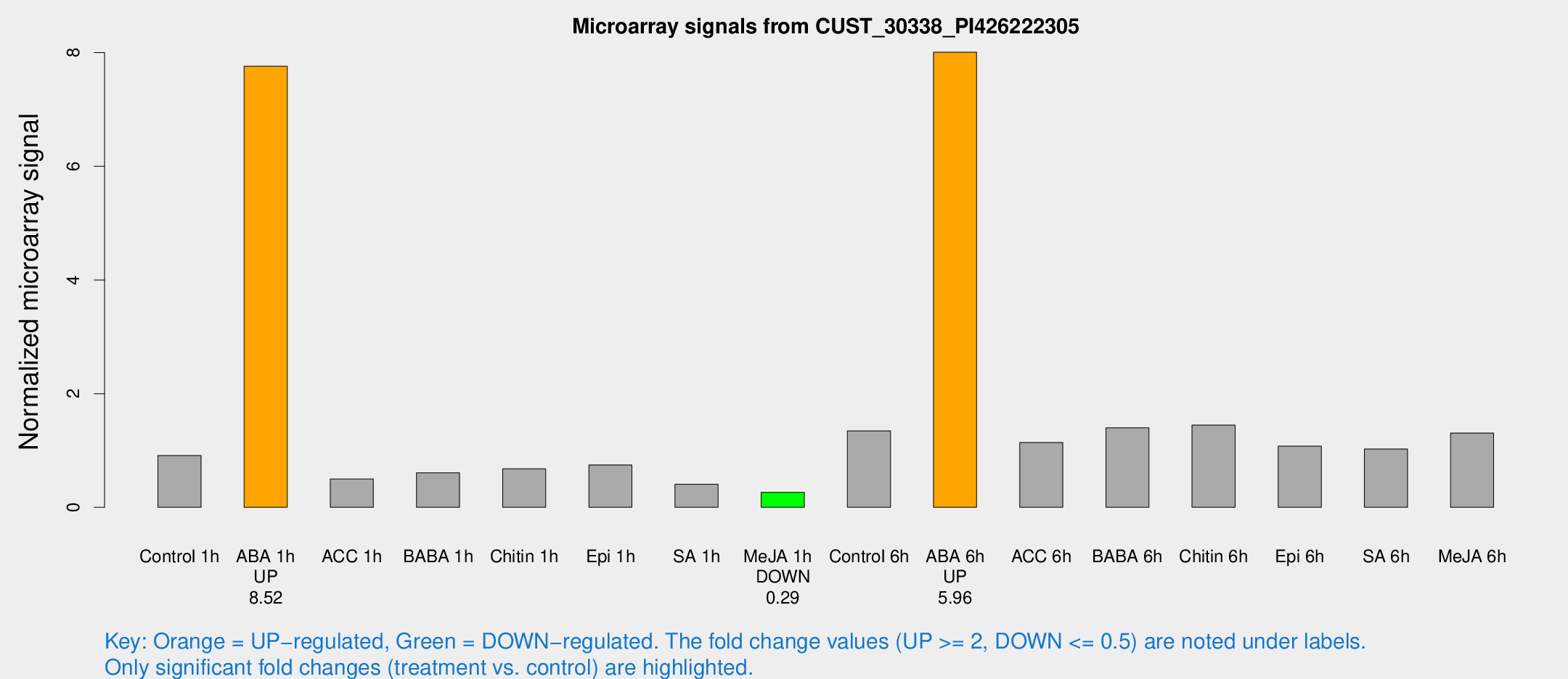
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 212.05 | 22.4028 | 0.910696 | 0.0575907 |
| ABA 1h | 1621.39 | 269.847 | 7.763 | 1.28032 |
| ACC 1h | 128.956 | 35.1237 | 0.498366 | 0.127468 |
| BABA 1h | 141.813 | 29.2087 | 0.607158 | 0.0814177 |
| Chitin 1h | 142.663 | 18.688 | 0.678718 | 0.0437567 |
| Epi 1h | 153.127 | 26.0386 | 0.745987 | 0.130931 |
| SA 1h | 104.906 | 29.0538 | 0.405936 | 0.106049 |
| Me-JA 1h | 49.7431 | 4.5232 | 0.263537 | 0.023938 |
| Control 6h | 313.757 | 35.3302 | 1.34421 | 0.11592 |
| ABA 6h | 2049.51 | 403.926 | 8.00728 | 1.40303 |
| ACC 6h | 305.758 | 38.0442 | 1.13922 | 0.107803 |
| BABA 6h | 376.447 | 74.8988 | 1.39751 | 0.267629 |
| Chitin 6h | 356.566 | 28.554 | 1.44653 | 0.140388 |
| Epi 6h | 283.395 | 34.9883 | 1.07563 | 0.193586 |
| SA 6h | 266.656 | 91.3017 | 1.02649 | 0.593234 |
| Me-JA 6h | 332.687 | 111.663 | 1.30493 | 0.330482 |
Source Transcript PGSC0003DMT400069946 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G11410.1 | +1 | 8e-106 | 126 | 73/112 (65%) | protein phosphatase 2CA | chr3:3584181-3585649 REVERSE LENGTH=399 |
