Probe CUST_30167_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30167_PI426222305 | JHI_St_60k_v1 | DMT400012257 | GGGTGTTGAATGGGTCGGGTTGGGTTAAGGATATTTTGAAAATAAAAGTGTTTGATTTAT |
All Microarray Probes Designed to Gene DMG400004810
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30167_PI426222305 | JHI_St_60k_v1 | DMT400012257 | GGGTGTTGAATGGGTCGGGTTGGGTTAAGGATATTTTGAAAATAAAAGTGTTTGATTTAT |
Microarray Signals from CUST_30167_PI426222305
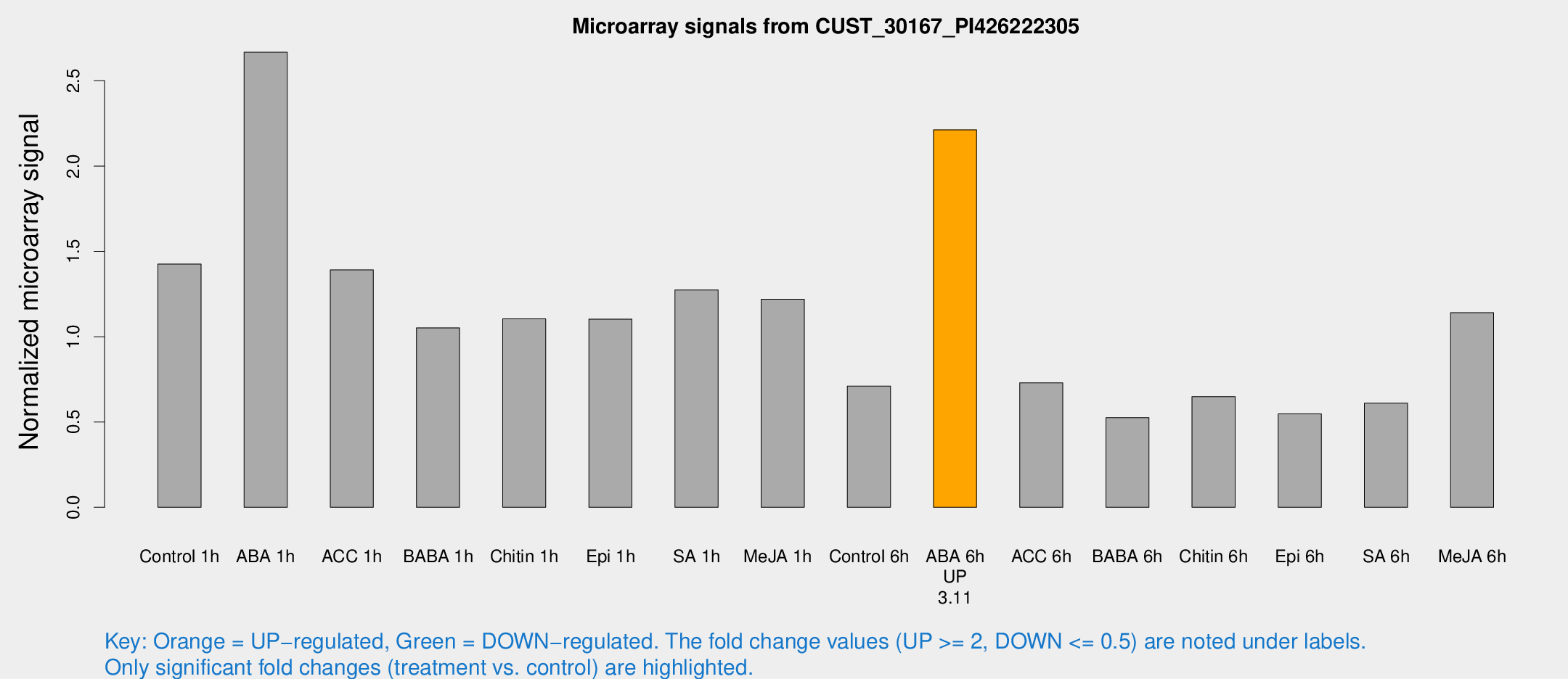
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 13410.4 | 1671.04 | 1.42541 | 0.0822968 |
| ABA 1h | 22136.8 | 2473.42 | 2.66734 | 0.294895 |
| ACC 1h | 14159.5 | 3357.09 | 1.39157 | 0.282053 |
| BABA 1h | 9920.38 | 2079.51 | 1.05197 | 0.146914 |
| Chitin 1h | 9392.35 | 1266.22 | 1.10487 | 0.0791818 |
| Epi 1h | 8893.95 | 644.435 | 1.10279 | 0.0781014 |
| SA 1h | 12720.1 | 2557.7 | 1.27357 | 0.23837 |
| Me-JA 1h | 9603.13 | 1753.74 | 1.21979 | 0.172973 |
| Control 6h | 6737.15 | 986.984 | 0.710351 | 0.0761767 |
| ABA 6h | 22110.6 | 2529.09 | 2.21246 | 0.159775 |
| ACC 6h | 7813.26 | 645.618 | 0.729529 | 0.168851 |
| BABA 6h | 5539.45 | 680.031 | 0.525259 | 0.0845212 |
| Chitin 6h | 6424.68 | 372.073 | 0.649278 | 0.0531252 |
| Epi 6h | 5784.1 | 545.684 | 0.548219 | 0.0316534 |
| SA 6h | 6700.67 | 2265.09 | 0.610111 | 0.235476 |
| Me-JA 6h | 10563.8 | 637.436 | 1.14039 | 0.0974413 |
Source Transcript PGSC0003DMT400012257 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G16563.1 | +3 | 0.0 | 558 | 301/505 (60%) | Eukaryotic aspartyl protease family protein | chr4:9329933-9331432 REVERSE LENGTH=499 |
