Probe CUST_30116_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30116_PI426222305 | JHI_St_60k_v1 | DMT400019543 | GCTAGATAATCATGGAAGGGTACCATGTAAGTTTTTGTTATCATGAACTATTGGTGTTCA |
All Microarray Probes Designed to Gene DMG400007556
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30116_PI426222305 | JHI_St_60k_v1 | DMT400019543 | GCTAGATAATCATGGAAGGGTACCATGTAAGTTTTTGTTATCATGAACTATTGGTGTTCA |
| CUST_30087_PI426222305 | JHI_St_60k_v1 | DMT400019544 | GCTAGATAATCATGGAAGGGTACCATGTAAGTTTTTGTTATCATGAACTATTGGTGTTCA |
Microarray Signals from CUST_30116_PI426222305
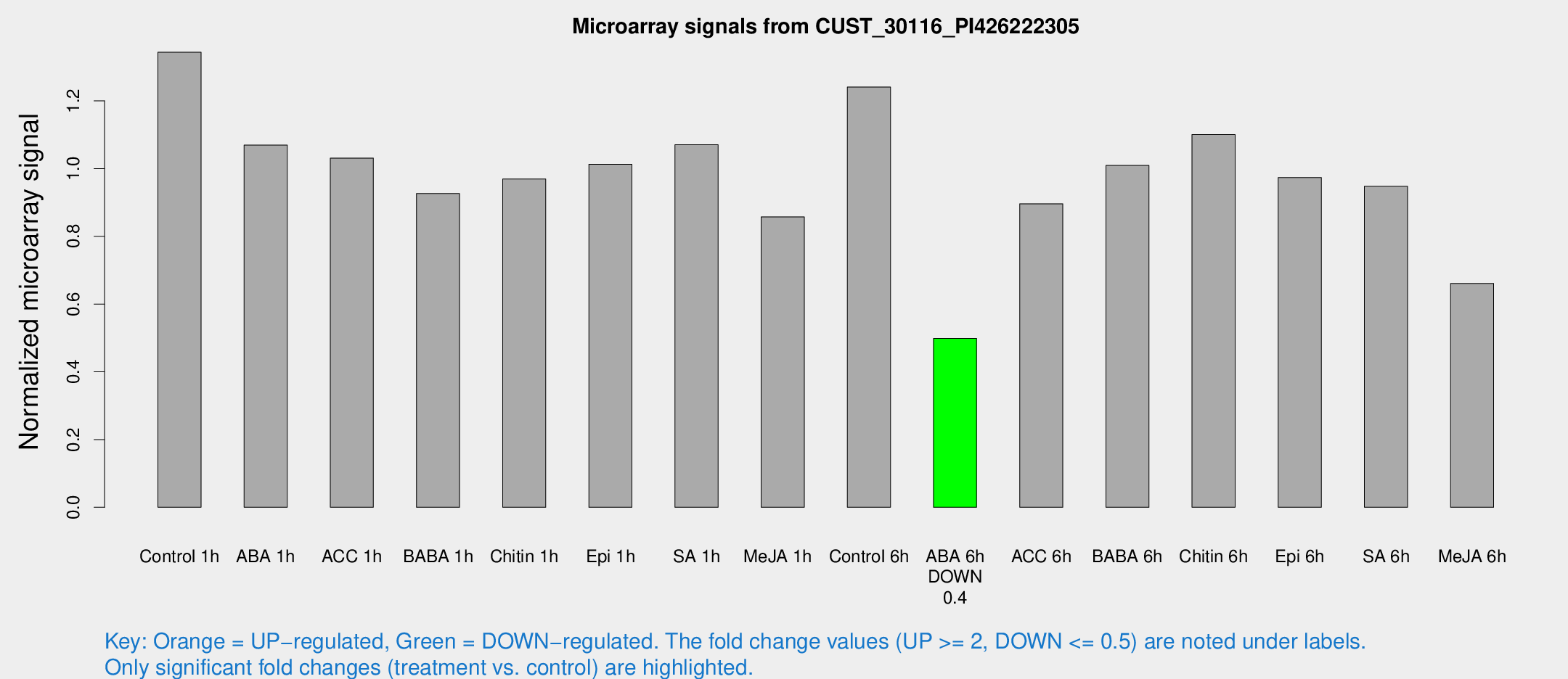
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5152.89 | 564.854 | 1.34354 | 0.0775748 |
| ABA 1h | 3587.41 | 207.47 | 1.06918 | 0.0617369 |
| ACC 1h | 4090.41 | 547.099 | 1.03102 | 0.0721952 |
| BABA 1h | 3633.84 | 860.754 | 0.926439 | 0.168028 |
| Chitin 1h | 3330.34 | 305.22 | 0.969237 | 0.0559678 |
| Epi 1h | 3321.54 | 191.921 | 1.01269 | 0.0584767 |
| SA 1h | 4257.9 | 647.936 | 1.07049 | 0.0946355 |
| Me-JA 1h | 2685.8 | 308.777 | 0.857497 | 0.0495205 |
| Control 6h | 4923.42 | 984.288 | 1.24094 | 0.173271 |
| ABA 6h | 2021.36 | 169.034 | 0.498664 | 0.0288047 |
| ACC 6h | 4028.14 | 703.896 | 0.896149 | 0.101347 |
| BABA 6h | 4352.59 | 584.336 | 1.00918 | 0.118578 |
| Chitin 6h | 4500.72 | 550.339 | 1.10023 | 0.113706 |
| Epi 6h | 4160.48 | 240.434 | 0.973551 | 0.0763562 |
| SA 6h | 3779.71 | 861.5 | 0.94777 | 0.141456 |
| Me-JA 6h | 2651.41 | 635.514 | 0.660807 | 0.113475 |
Source Transcript PGSC0003DMT400019543 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G16910.1 | +3 | 2e-162 | 472 | 225/299 (75%) | acyl-activating enzyme 7 | chr3:5773231-5775411 REVERSE LENGTH=569 |
