Probe CUST_30087_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30087_PI426222305 | JHI_St_60k_v1 | DMT400019544 | GCTAGATAATCATGGAAGGGTACCATGTAAGTTTTTGTTATCATGAACTATTGGTGTTCA |
All Microarray Probes Designed to Gene DMG400007556
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30087_PI426222305 | JHI_St_60k_v1 | DMT400019544 | GCTAGATAATCATGGAAGGGTACCATGTAAGTTTTTGTTATCATGAACTATTGGTGTTCA |
| CUST_30116_PI426222305 | JHI_St_60k_v1 | DMT400019543 | GCTAGATAATCATGGAAGGGTACCATGTAAGTTTTTGTTATCATGAACTATTGGTGTTCA |
Microarray Signals from CUST_30087_PI426222305
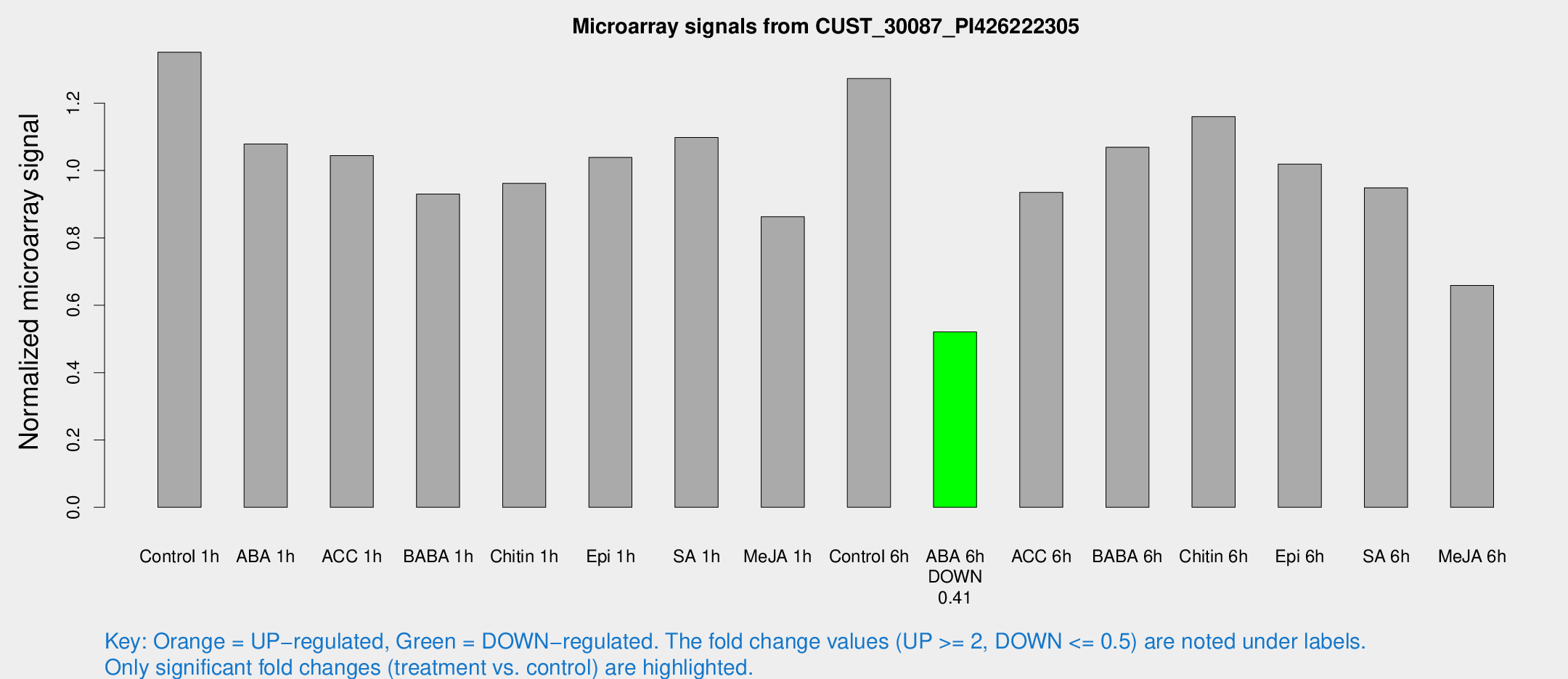
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5012.01 | 481.361 | 1.3514 | 0.078029 |
| ABA 1h | 3518.33 | 203.73 | 1.07901 | 0.0623053 |
| ACC 1h | 4012.65 | 506.189 | 1.04431 | 0.0615899 |
| BABA 1h | 3488.58 | 749.174 | 0.930018 | 0.136302 |
| Chitin 1h | 3191.64 | 198.507 | 0.961869 | 0.0555435 |
| Epi 1h | 3313.32 | 191.76 | 1.03896 | 0.059994 |
| SA 1h | 4214.37 | 521.821 | 1.09843 | 0.0719273 |
| Me-JA 1h | 2599.79 | 165.522 | 0.862789 | 0.0498273 |
| Control 6h | 4942 | 1092.82 | 1.27328 | 0.208839 |
| ABA 6h | 2044.52 | 139.194 | 0.520778 | 0.0300844 |
| ACC 6h | 4103.42 | 786.7 | 0.934967 | 0.104058 |
| BABA 6h | 4449.58 | 481.984 | 1.06902 | 0.0973899 |
| Chitin 6h | 4634.6 | 660.243 | 1.16014 | 0.154486 |
| Epi 6h | 4229.67 | 244.574 | 1.01913 | 0.0829614 |
| SA 6h | 3687.36 | 862.564 | 0.948649 | 0.150137 |
| Me-JA 6h | 2569.16 | 624.175 | 0.658838 | 0.114714 |
Source Transcript PGSC0003DMT400019544 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G16910.1 | +2 | 0.0 | 877 | 412/563 (73%) | acyl-activating enzyme 7 | chr3:5773231-5775411 REVERSE LENGTH=569 |
