Probe CUST_30036_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30036_PI426222305 | JHI_St_60k_v1 | DMT400035910 | AAGGAGAAAGAAGAGGATTTCATGGCTATCAAAGGCTCAAAGTTGCCTCAAAGACCCAAA |
All Microarray Probes Designed to Gene DMG400013825
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30036_PI426222305 | JHI_St_60k_v1 | DMT400035910 | AAGGAGAAAGAAGAGGATTTCATGGCTATCAAAGGCTCAAAGTTGCCTCAAAGACCCAAA |
Microarray Signals from CUST_30036_PI426222305
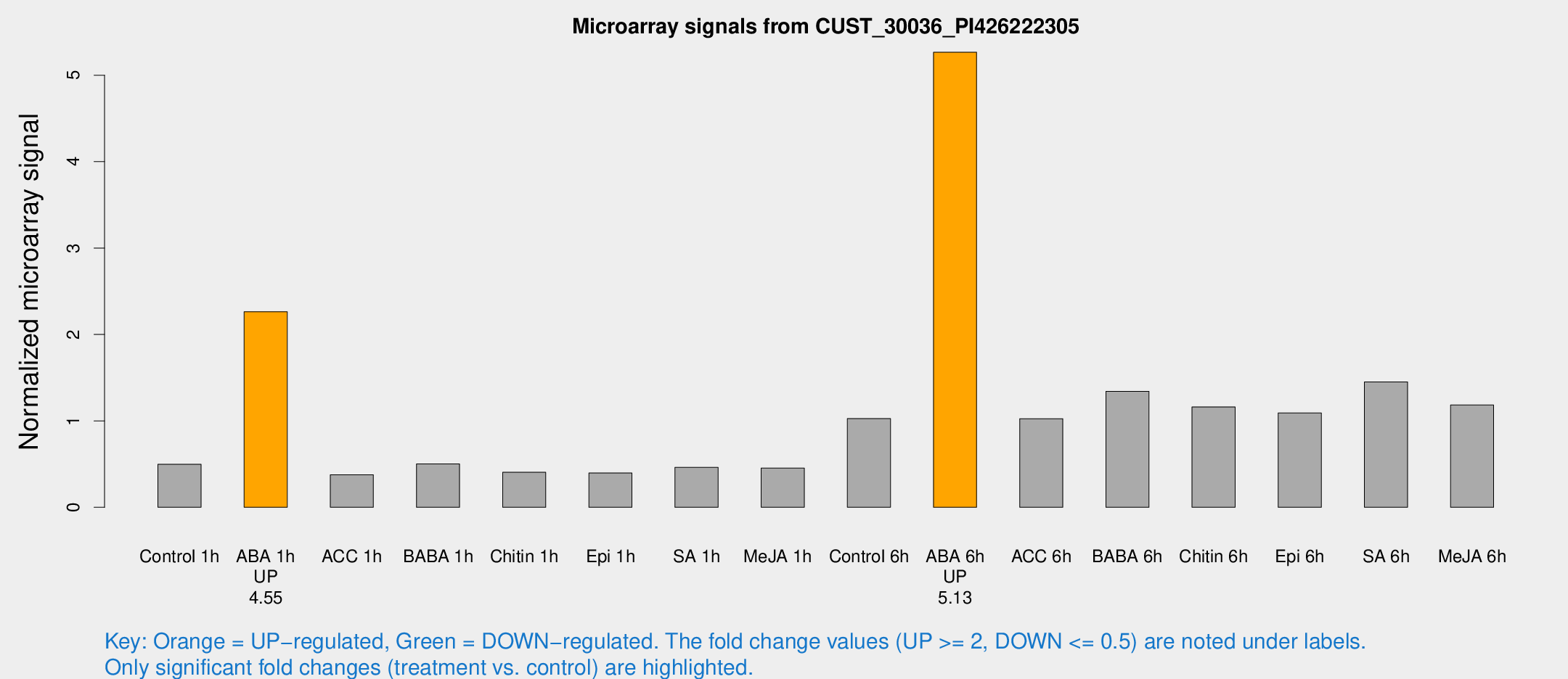
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 597.414 | 54.8711 | 0.497162 | 0.0465811 |
| ABA 1h | 2453.71 | 390.996 | 2.26367 | 0.249248 |
| ACC 1h | 504.02 | 138.864 | 0.376425 | 0.095705 |
| BABA 1h | 593.107 | 95.4293 | 0.501832 | 0.038084 |
| Chitin 1h | 479.433 | 143.506 | 0.404817 | 0.131293 |
| Epi 1h | 451.322 | 122.284 | 0.396512 | 0.138556 |
| SA 1h | 587.493 | 121.29 | 0.461724 | 0.0658256 |
| Me-JA 1h | 441.71 | 25.7554 | 0.453149 | 0.0370647 |
| Control 6h | 1369.64 | 400.96 | 1.02576 | 0.290115 |
| ABA 6h | 6704.92 | 491.303 | 5.26563 | 0.304022 |
| ACC 6h | 1442.63 | 231.987 | 1.02537 | 0.117114 |
| BABA 6h | 1792.03 | 103.626 | 1.34255 | 0.0775574 |
| Chitin 6h | 1474.75 | 85.3324 | 1.1614 | 0.0777432 |
| Epi 6h | 1475.67 | 119.798 | 1.09099 | 0.0630465 |
| SA 6h | 1708.49 | 98.7011 | 1.44991 | 0.180463 |
| Me-JA 6h | 1478.82 | 349.747 | 1.18324 | 0.178798 |
Source Transcript PGSC0003DMT400035910 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G55340.1 | +2 | 1e-38 | 136 | 71/101 (70%) | Protein of unknown function (DUF1639) | chr1:20652605-20653469 FORWARD LENGTH=205 |
