Probe CUST_3001_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3001_PI426222305 | JHI_St_60k_v1 | DMT400033215 | AGTCCACAGGCCACAAACATCTAAAACAGCCGAGATACATGTGCTCAAGCTAAGAAGAAG |
All Microarray Probes Designed to Gene DMG400012753
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2797_PI426222305 | JHI_St_60k_v1 | DMT400033216 | GCAAAAGAGCTCTTCTTCTTCATCAAGAAAACAAAGCCATGTTCACTCAAATTATTCAAC |
| CUST_3001_PI426222305 | JHI_St_60k_v1 | DMT400033215 | AGTCCACAGGCCACAAACATCTAAAACAGCCGAGATACATGTGCTCAAGCTAAGAAGAAG |
Microarray Signals from CUST_3001_PI426222305
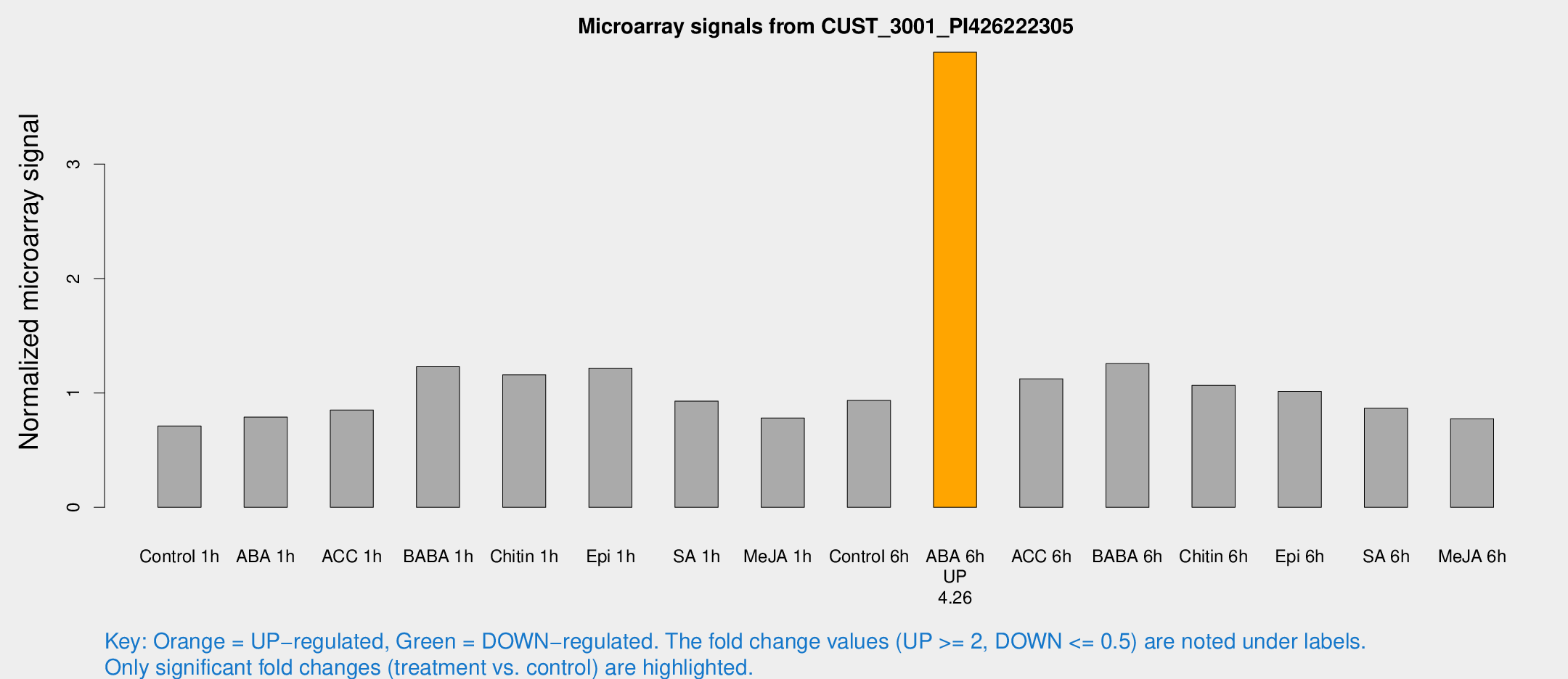
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 29.1748 | 6.25319 | 0.710798 | 0.188959 |
| ABA 1h | 28.7725 | 7.53618 | 0.788205 | 0.254482 |
| ACC 1h | 35.5197 | 7.05903 | 0.850085 | 0.144562 |
| BABA 1h | 46.658 | 6.09825 | 1.22844 | 0.135065 |
| Chitin 1h | 41.5922 | 6.98912 | 1.15843 | 0.181107 |
| Epi 1h | 40.8969 | 4.03739 | 1.21632 | 0.120872 |
| SA 1h | 39.037 | 8.23175 | 0.928596 | 0.254785 |
| Me-JA 1h | 27.9132 | 9.87122 | 0.781149 | 0.387982 |
| Control 6h | 42.4264 | 15.1733 | 0.933994 | 0.340615 |
| ABA 6h | 165.084 | 13.1535 | 3.97833 | 0.245397 |
| ACC 6h | 51.181 | 7.48299 | 1.12346 | 0.117024 |
| BABA 6h | 54.9666 | 5.0367 | 1.25593 | 0.114295 |
| Chitin 6h | 44.0564 | 4.57533 | 1.06652 | 0.110829 |
| Epi 6h | 45.9133 | 8.81002 | 1.01312 | 0.263101 |
| SA 6h | 33.6797 | 4.16149 | 0.866483 | 0.198063 |
| Me-JA 6h | 32.2803 | 7.88136 | 0.774127 | 0.171539 |
Source Transcript PGSC0003DMT400033215 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
