Probe CUST_29986_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29986_PI426222305 | JHI_St_60k_v1 | DMT400065485 | GGAGTTCTGTCTGCTGCATCACCTCTTCGATTTTATTTATTGCAACTTACATTATAAATC |
All Microarray Probes Designed to Gene DMG400025467
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29986_PI426222305 | JHI_St_60k_v1 | DMT400065485 | GGAGTTCTGTCTGCTGCATCACCTCTTCGATTTTATTTATTGCAACTTACATTATAAATC |
Microarray Signals from CUST_29986_PI426222305
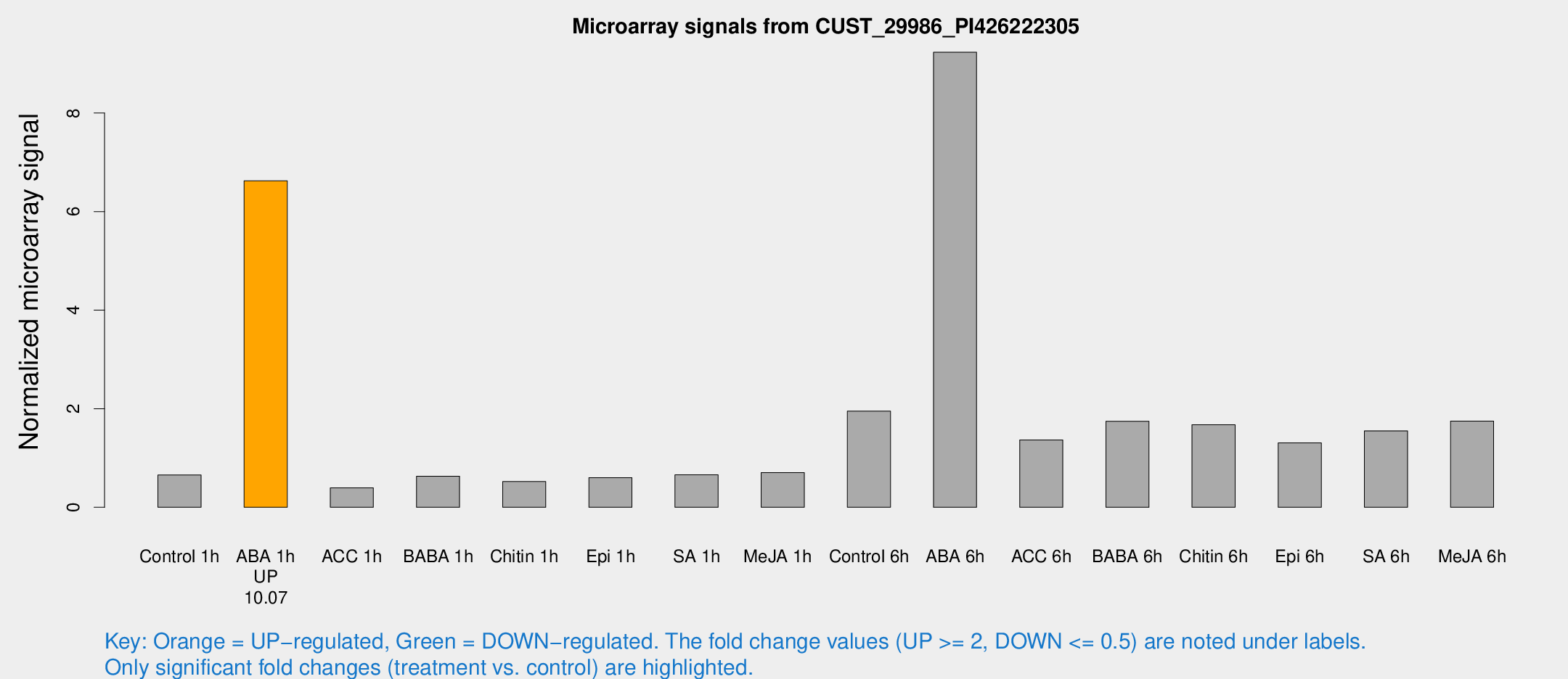
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 87.6588 | 6.07508 | 0.657701 | 0.0456206 |
| ABA 1h | 807.63 | 147.927 | 6.62398 | 1.11153 |
| ACC 1h | 55.9825 | 11.7014 | 0.393526 | 0.0683883 |
| BABA 1h | 82.6053 | 11.3861 | 0.631378 | 0.046702 |
| Chitin 1h | 62.9106 | 4.96767 | 0.523234 | 0.0412259 |
| Epi 1h | 71.1441 | 10.5482 | 0.602417 | 0.0898872 |
| SA 1h | 93.2561 | 16.255 | 0.661098 | 0.14911 |
| Me-JA 1h | 78.5937 | 12.9169 | 0.705562 | 0.0678471 |
| Control 6h | 278.163 | 72.2931 | 1.95193 | 0.402482 |
| ABA 6h | 1399.86 | 330.021 | 9.2347 | 2.1756 |
| ACC 6h | 219.519 | 44.7694 | 1.36736 | 0.27844 |
| BABA 6h | 280.103 | 66.9558 | 1.74708 | 0.454569 |
| Chitin 6h | 245.452 | 42.6636 | 1.6765 | 0.340041 |
| Epi 6h | 203.112 | 35.6003 | 1.30596 | 0.35518 |
| SA 6h | 240.249 | 84.2393 | 1.55147 | 1.02126 |
| Me-JA 6h | 269.873 | 97.5213 | 1.7507 | 0.602163 |
Source Transcript PGSC0003DMT400065485 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
