Probe CUST_29844_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29844_PI426222305 | JHI_St_60k_v1 | DMT400033417 | CTTGTAATGAACATTGGTGGTTGCTTGTGTTACAACCCAATCTTCATATAGTAATACATG |
All Microarray Probes Designed to Gene DMG400012839
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29844_PI426222305 | JHI_St_60k_v1 | DMT400033417 | CTTGTAATGAACATTGGTGGTTGCTTGTGTTACAACCCAATCTTCATATAGTAATACATG |
Microarray Signals from CUST_29844_PI426222305
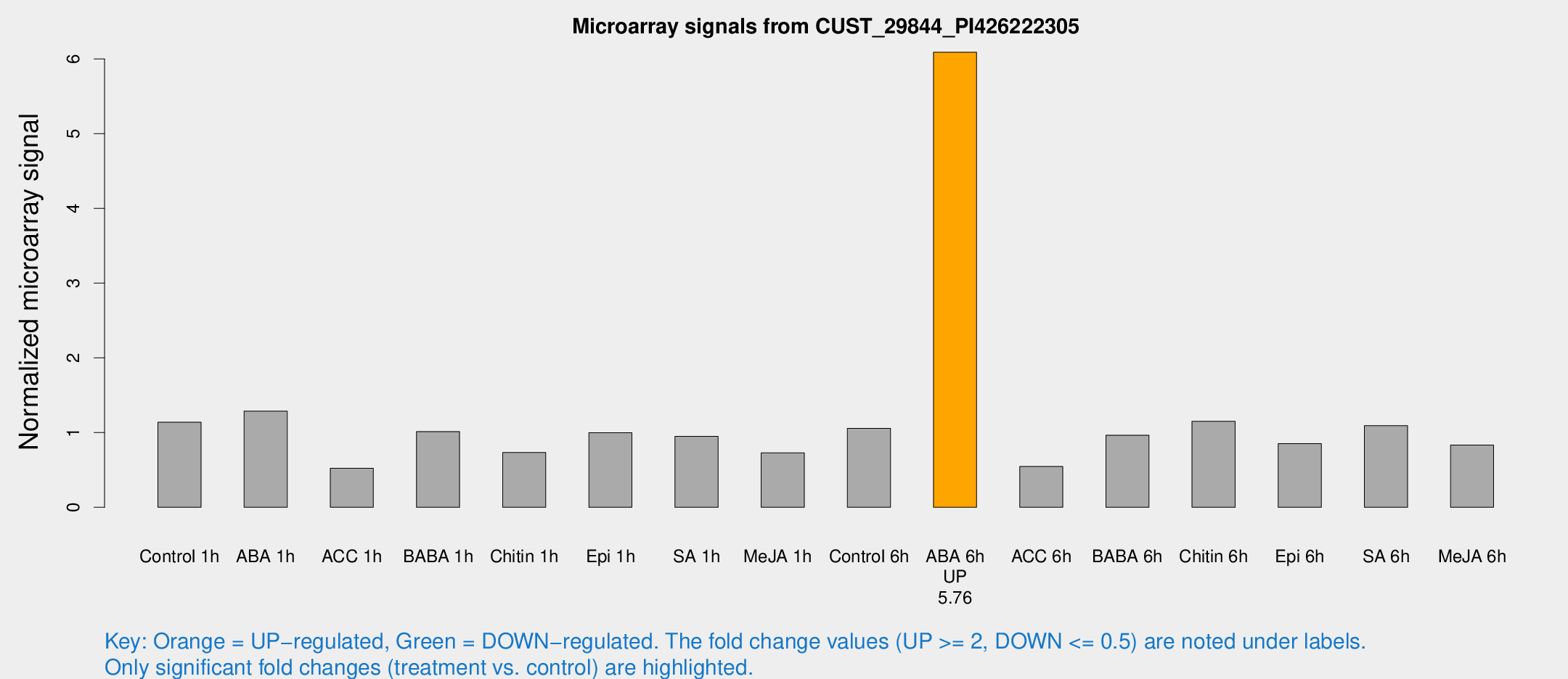
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 32332.3 | 1866.77 | 1.13845 | 0.0825019 |
| ABA 1h | 34095.2 | 7440.37 | 1.2883 | 0.267861 |
| ACC 1h | 21289 | 8754.8 | 0.521805 | 0.415723 |
| BABA 1h | 27839.2 | 1611.71 | 1.01416 | 0.159053 |
| Chitin 1h | 20497.1 | 5338.64 | 0.7338 | 0.255605 |
| Epi 1h | 26672.9 | 7182.13 | 0.998658 | 0.309519 |
| SA 1h | 28279.4 | 3770.39 | 0.950255 | 0.198327 |
| Me-JA 1h | 17424.3 | 2900.4 | 0.728867 | 0.131087 |
| Control 6h | 31331.1 | 6017.9 | 1.05666 | 0.120653 |
| ABA 6h | 185908 | 18617.2 | 6.09091 | 0.376676 |
| ACC 6h | 18527.7 | 3510.7 | 0.546811 | 0.138411 |
| BABA 6h | 30957.6 | 2821.14 | 0.965131 | 0.0576771 |
| Chitin 6h | 34907.2 | 2021.5 | 1.15121 | 0.0664654 |
| Epi 6h | 27735.7 | 3443.5 | 0.8531 | 0.160642 |
| SA 6h | 30720.9 | 1775.67 | 1.09236 | 0.137263 |
| Me-JA 6h | 24961.2 | 6227.27 | 0.834084 | 0.140792 |
Source Transcript PGSC0003DMT400033417 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38540.1 | +3 | 2e-24 | 96 | 58/113 (51%) | lipid transfer protein 1 | chr2:16130418-16130893 FORWARD LENGTH=118 |
