Probe CUST_29771_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29771_PI426222305 | JHI_St_60k_v1 | DMT400053837 | TTTTGACCTGGCTTGTGTTCTGCCAGTATAGGTACTTATAACAGCAAACCGAGCGGTTTG |
All Microarray Probes Designed to Gene DMG400020882
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29771_PI426222305 | JHI_St_60k_v1 | DMT400053837 | TTTTGACCTGGCTTGTGTTCTGCCAGTATAGGTACTTATAACAGCAAACCGAGCGGTTTG |
Microarray Signals from CUST_29771_PI426222305
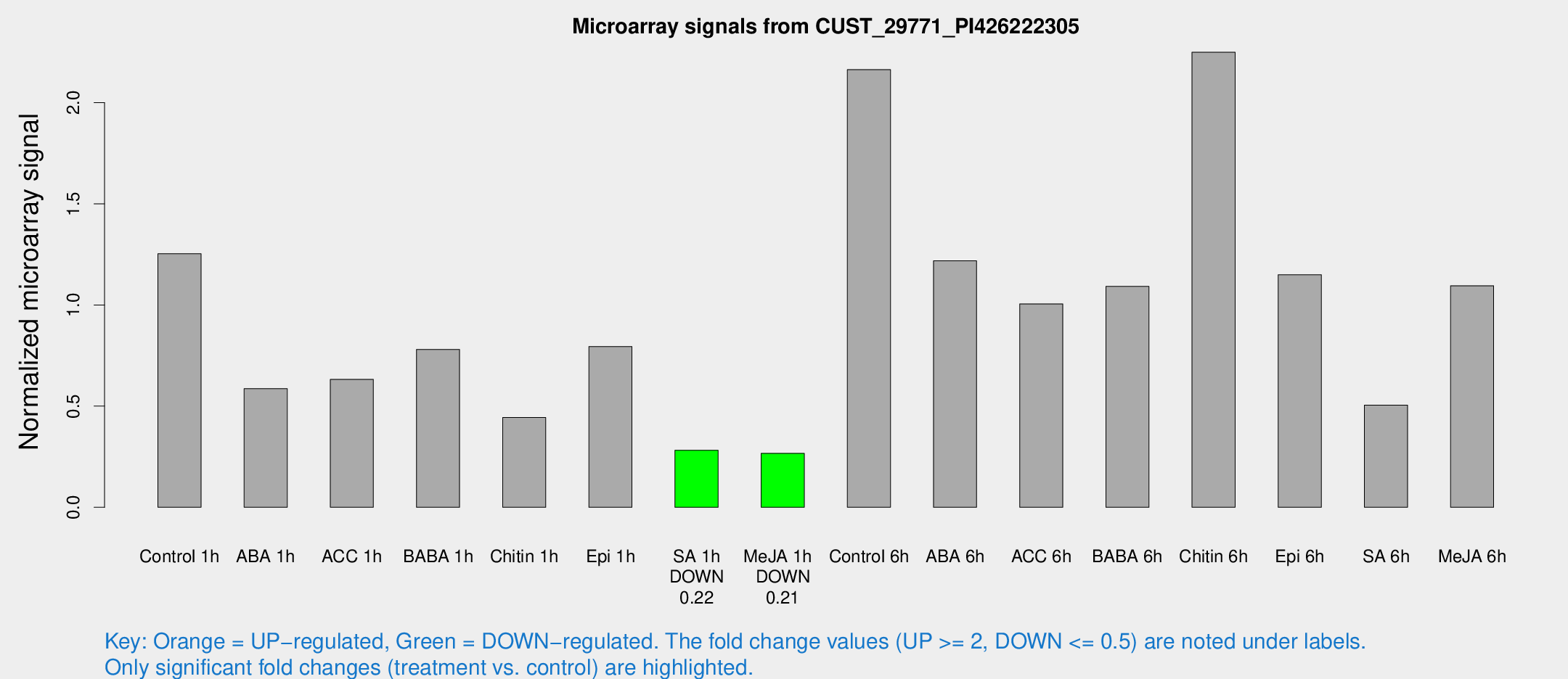
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 35.2913 | 4.62887 | 1.25355 | 0.15705 |
| ABA 1h | 16.901 | 5.73957 | 0.586528 | 0.351694 |
| ACC 1h | 19.8782 | 5.73605 | 0.632411 | 0.207964 |
| BABA 1h | 22.8943 | 6.58992 | 0.780554 | 0.180934 |
| Chitin 1h | 13.5652 | 6.17167 | 0.443996 | 0.21946 |
| Epi 1h | 21.0756 | 6.05361 | 0.794816 | 0.31646 |
| SA 1h | 8.04848 | 3.43964 | 0.281955 | 0.121363 |
| Me-JA 1h | 6.04172 | 3.50992 | 0.266692 | 0.154604 |
| Control 6h | 63.1115 | 13.0865 | 2.16349 | 0.322672 |
| ABA 6h | 36.2205 | 4.21112 | 1.21798 | 0.145097 |
| ACC 6h | 32.1282 | 4.72299 | 1.0053 | 0.144544 |
| BABA 6h | 40.7388 | 16.7502 | 1.09188 | 0.51813 |
| Chitin 6h | 71.674 | 17.6027 | 2.24922 | 0.707285 |
| Epi 6h | 46.2098 | 18.8321 | 1.14899 | 0.950705 |
| SA 6h | 22.3613 | 15.2046 | 0.50519 | 0.693589 |
| Me-JA 6h | 38.0469 | 18.8114 | 1.09504 | 0.482884 |
Source Transcript PGSC0003DMT400053837 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | ATMG00070.1 | +2 | 1e-37 | 129 | 63/75 (84%) | NADH dehydrogenase subunit 9 | chrM:23663-24235 REVERSE LENGTH=190 |
