Probe CUST_29514_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29514_PI426222305 | JHI_St_60k_v1 | DMT400075879 | ATTGTTGCATTTGTAGAGGAGGCAGGGGTCTTCAATTTCCAAATGAGTAAAATGTTAGGA |
All Microarray Probes Designed to Gene DMG400029503
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29514_PI426222305 | JHI_St_60k_v1 | DMT400075879 | ATTGTTGCATTTGTAGAGGAGGCAGGGGTCTTCAATTTCCAAATGAGTAAAATGTTAGGA |
| CUST_29503_PI426222305 | JHI_St_60k_v1 | DMT400075880 | ATTGTTGCATTTGTAGAGGAGGCAGGGGTCTTCAATTTCCAAATGAGTAAAATGTTAGGA |
Microarray Signals from CUST_29514_PI426222305
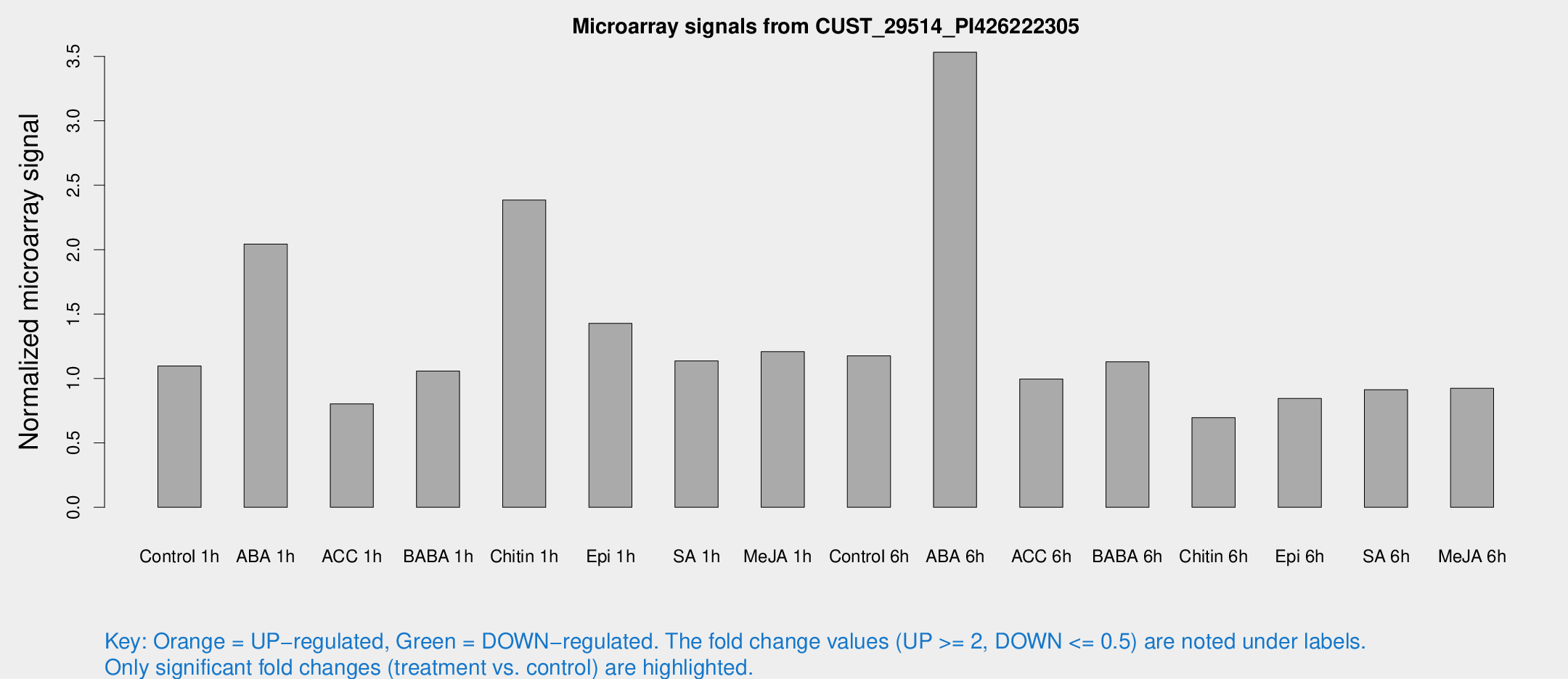
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 12.7468 | 5.48051 | 1.09671 | 0.458806 |
| ABA 1h | 20.7207 | 6.84257 | 2.04354 | 1.0547 |
| ACC 1h | 8.51217 | 4.34931 | 0.803282 | 0.424482 |
| BABA 1h | 10.3152 | 3.91218 | 1.05698 | 0.436226 |
| Chitin 1h | 23.9854 | 8.55669 | 2.38494 | 1.10387 |
| Epi 1h | 13.3228 | 4.12024 | 1.42759 | 0.511314 |
| SA 1h | 15.6072 | 8.86993 | 1.13702 | 0.670503 |
| Me-JA 1h | 10.7674 | 3.82355 | 1.20771 | 0.517263 |
| Control 6h | 13.4732 | 5.45497 | 1.17634 | 0.471376 |
| ABA 6h | 38.4529 | 7.76326 | 3.53244 | 0.679113 |
| ACC 6h | 11.4697 | 4.80222 | 0.99505 | 0.416279 |
| BABA 6h | 14.3777 | 5.62184 | 1.12961 | 0.450085 |
| Chitin 6h | 7.27883 | 4.22691 | 0.696317 | 0.403908 |
| Epi 6h | 9.60207 | 4.85872 | 0.84443 | 0.434509 |
| SA 6h | 9.21143 | 4.14754 | 0.913236 | 0.449113 |
| Me-JA 6h | 9.42811 | 3.84167 | 0.923527 | 0.411826 |
Source Transcript PGSC0003DMT400075879 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G14720.1 | +3 | 2e-66 | 218 | 99/146 (68%) | xyloglucan endotransglucosylase/hydrolase 28 | chr1:5066806-5068466 REVERSE LENGTH=332 |
