Probe CUST_29503_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29503_PI426222305 | JHI_St_60k_v1 | DMT400075880 | ATTGTTGCATTTGTAGAGGAGGCAGGGGTCTTCAATTTCCAAATGAGTAAAATGTTAGGA |
All Microarray Probes Designed to Gene DMG400029503
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29503_PI426222305 | JHI_St_60k_v1 | DMT400075880 | ATTGTTGCATTTGTAGAGGAGGCAGGGGTCTTCAATTTCCAAATGAGTAAAATGTTAGGA |
| CUST_29514_PI426222305 | JHI_St_60k_v1 | DMT400075879 | ATTGTTGCATTTGTAGAGGAGGCAGGGGTCTTCAATTTCCAAATGAGTAAAATGTTAGGA |
Microarray Signals from CUST_29503_PI426222305
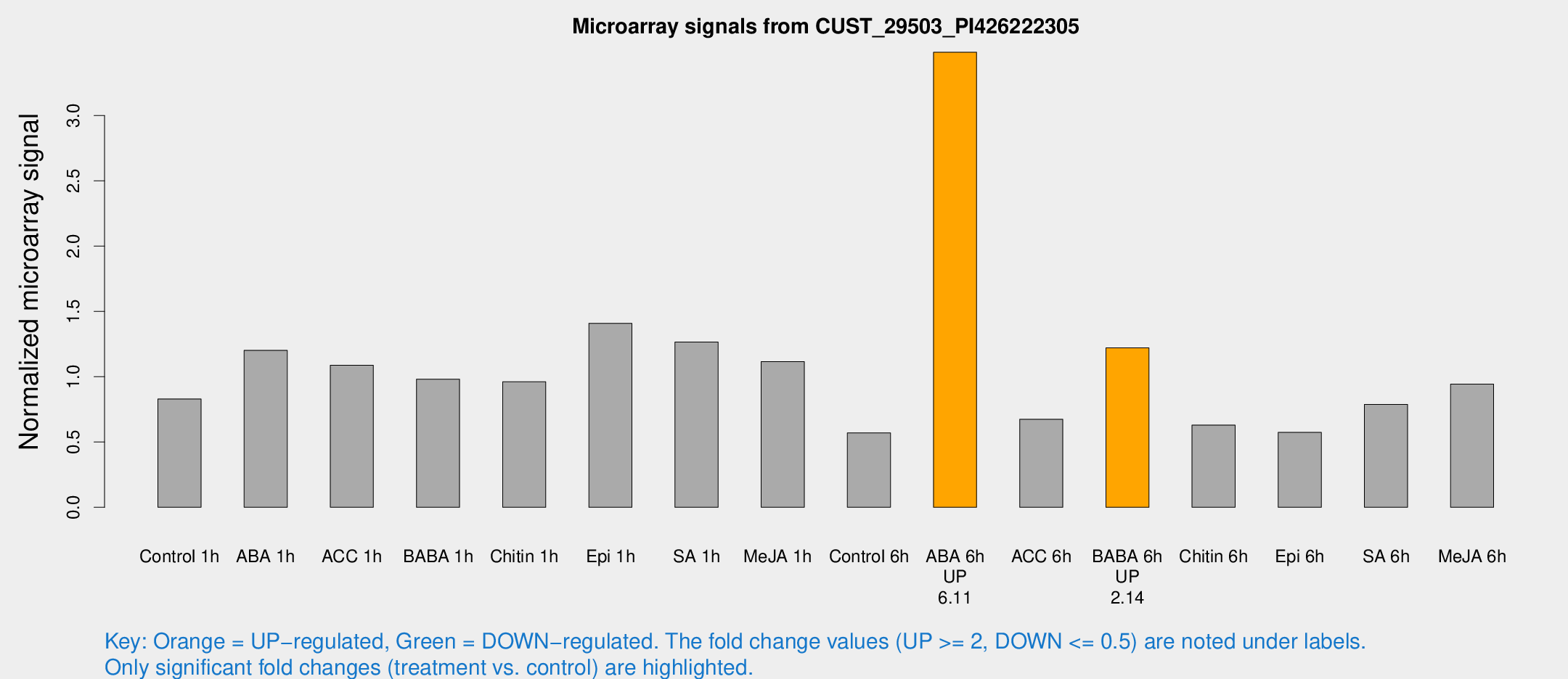
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.46464 | 3.27112 | 0.830199 | 0.324495 |
| ABA 1h | 12.9143 | 3.83697 | 1.20228 | 0.460868 |
| ACC 1h | 14.2309 | 5.3989 | 1.08732 | 0.62934 |
| BABA 1h | 10.4994 | 3.52874 | 0.981161 | 0.349681 |
| Chitin 1h | 9.81053 | 3.37459 | 0.961045 | 0.374924 |
| Epi 1h | 13.3456 | 3.36918 | 1.4092 | 0.364909 |
| SA 1h | 17.5807 | 7.59855 | 1.26581 | 0.712879 |
| Me-JA 1h | 10.5679 | 3.43702 | 1.11516 | 0.429289 |
| Control 6h | 6.19036 | 3.49199 | 0.570339 | 0.321943 |
| ABA 6h | 41.5922 | 8.27909 | 3.48531 | 0.480322 |
| ACC 6h | 8.62034 | 4.22981 | 0.674778 | 0.337097 |
| BABA 6h | 14.9406 | 3.99983 | 1.22103 | 0.329721 |
| Chitin 6h | 7.29583 | 3.81287 | 0.629769 | 0.332613 |
| Epi 6h | 7.11621 | 3.99682 | 0.573942 | 0.322273 |
| SA 6h | 8.85395 | 3.68065 | 0.787657 | 0.364439 |
| Me-JA 6h | 11.0113 | 3.56045 | 0.943711 | 0.363261 |
Source Transcript PGSC0003DMT400075880 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G14720.1 | +1 | 2e-155 | 448 | 210/311 (68%) | xyloglucan endotransglucosylase/hydrolase 28 | chr1:5066806-5068466 REVERSE LENGTH=332 |
