Probe CUST_29464_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29464_PI426222305 | JHI_St_60k_v1 | DMT400075698 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
All Microarray Probes Designed to Gene DMG401029444
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29494_PI426222305 | JHI_St_60k_v1 | DMT400075694 | AGAGTAGTTGGTACTTATGGATACATGGCCCCTGAATATGCAATGAAAGGAAGATTCTCT |
| CUST_29464_PI426222305 | JHI_St_60k_v1 | DMT400075698 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
| CUST_29484_PI426222305 | JHI_St_60k_v1 | DMT400075702 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
| CUST_29415_PI426222305 | JHI_St_60k_v1 | DMT400075696 | TAAAGAGGTCTGACCGACTATATCATGTACCTTATATTTCAGGCATGGAAGTTATGGAAA |
| CUST_29383_PI426222305 | JHI_St_60k_v1 | DMT400075701 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
| CUST_29468_PI426222305 | JHI_St_60k_v1 | DMT400075700 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
| CUST_29461_PI426222305 | JHI_St_60k_v1 | DMT400075699 | GCACCATCACAACCTGCTTTTACTGAAAGACATGATTGTATCTTCAAAATGTGCAATGTA |
Microarray Signals from CUST_29464_PI426222305
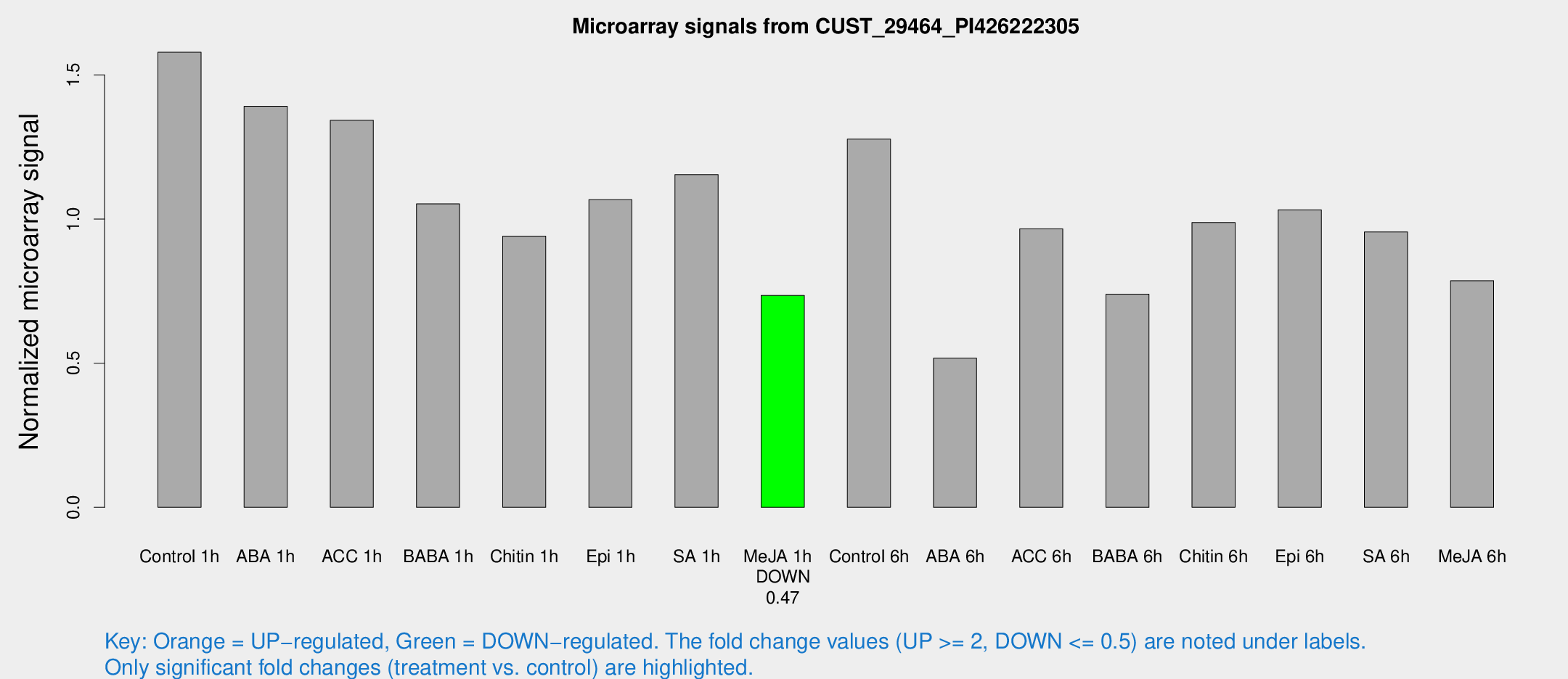
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 873.334 | 104.59 | 1.57877 | 0.0913519 |
| ABA 1h | 672.223 | 39.002 | 1.39129 | 0.119257 |
| ACC 1h | 807.976 | 191.047 | 1.34305 | 0.282417 |
| BABA 1h | 576.47 | 106.983 | 1.0527 | 0.110837 |
| Chitin 1h | 465.673 | 46.929 | 0.940624 | 0.107745 |
| Epi 1h | 504.387 | 29.3381 | 1.06716 | 0.061988 |
| SA 1h | 647.319 | 37.5645 | 1.15381 | 0.069238 |
| Me-JA 1h | 328.448 | 20.6817 | 0.735013 | 0.0430624 |
| Control 6h | 747.59 | 174.934 | 1.27718 | 0.234957 |
| ABA 6h | 305.641 | 43.986 | 0.517123 | 0.0503437 |
| ACC 6h | 614.408 | 77.5683 | 0.96621 | 0.0606749 |
| BABA 6h | 455.089 | 41.9176 | 0.739558 | 0.0602785 |
| Chitin 6h | 576.071 | 39.2352 | 0.987712 | 0.0804748 |
| Epi 6h | 640.36 | 62.4907 | 1.03177 | 0.150243 |
| SA 6h | 534.917 | 98.5364 | 0.955415 | 0.0862878 |
| Me-JA 6h | 442.408 | 84.029 | 0.786109 | 0.0839875 |
Source Transcript PGSC0003DMT400075698 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G11330.2 | +1 | 8e-85 | 275 | 136/226 (60%) | S-locus lectin protein kinase family protein | chr1:3810372-3813416 FORWARD LENGTH=842 |
