Probe CUST_29450_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29450_PI426222305 | JHI_St_60k_v1 | DMT400075850 | ATCGCGCATATCACAAGATAGACTTCGTTTCTTTCTAGCTCTTTCTTCTTTGAATTTCAC |
All Microarray Probes Designed to Gene DMG400029490
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29450_PI426222305 | JHI_St_60k_v1 | DMT400075850 | ATCGCGCATATCACAAGATAGACTTCGTTTCTTTCTAGCTCTTTCTTCTTTGAATTTCAC |
Microarray Signals from CUST_29450_PI426222305
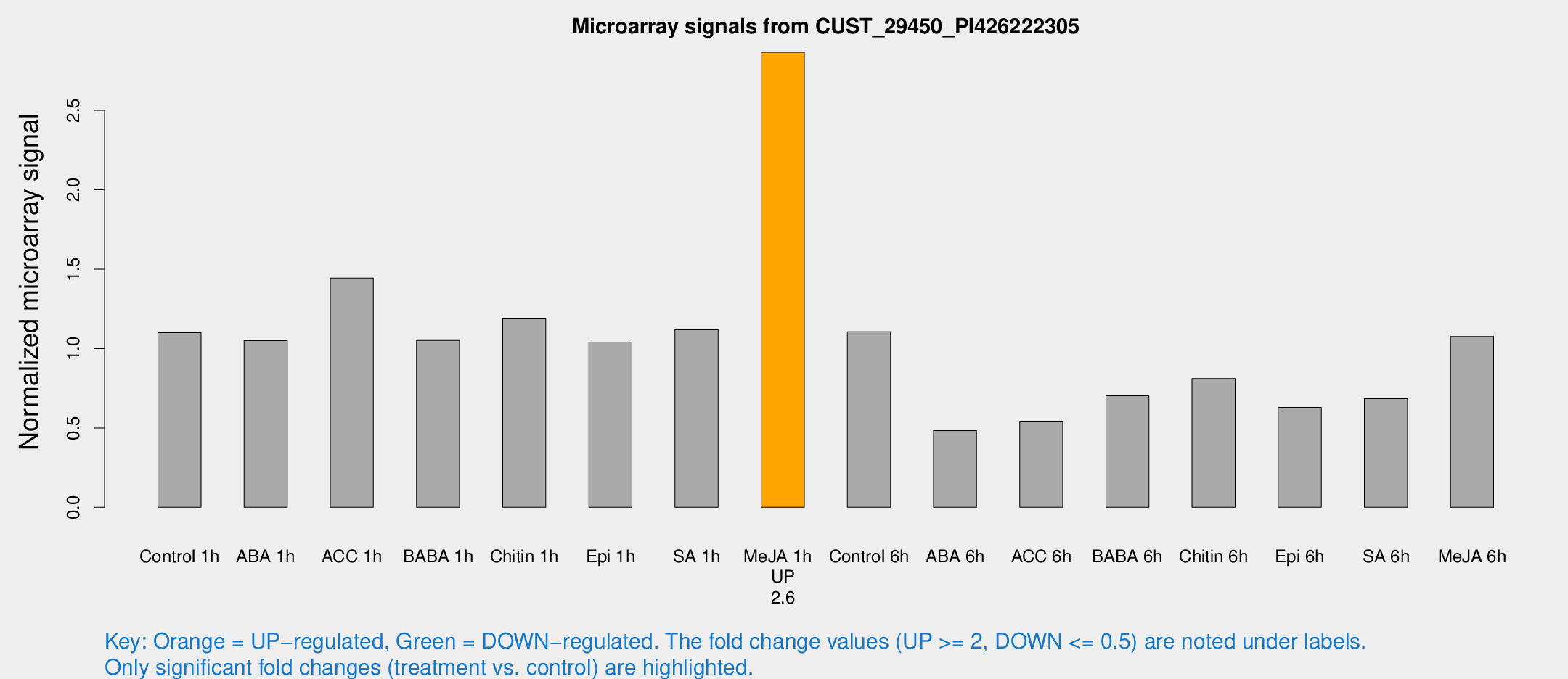
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1103.25 | 138.705 | 1.10056 | 0.0846049 |
| ABA 1h | 933.564 | 125.021 | 1.05054 | 0.0918244 |
| ACC 1h | 1471.51 | 102.555 | 1.4435 | 0.0834271 |
| BABA 1h | 1019.06 | 129.566 | 1.05163 | 0.0608449 |
| Chitin 1h | 1090.61 | 192.706 | 1.18681 | 0.18022 |
| Epi 1h | 901.397 | 103.758 | 1.04117 | 0.0894159 |
| SA 1h | 1143.4 | 98.1551 | 1.1182 | 0.0646507 |
| Me-JA 1h | 2316.93 | 134.171 | 2.86605 | 0.31129 |
| Control 6h | 1203.17 | 354.073 | 1.10596 | 0.25705 |
| ABA 6h | 521.171 | 80.7421 | 0.484052 | 0.0702845 |
| ACC 6h | 628.123 | 105.477 | 0.538111 | 0.045254 |
| BABA 6h | 779.791 | 48.0012 | 0.702734 | 0.0420325 |
| Chitin 6h | 864.193 | 102.14 | 0.811122 | 0.0922842 |
| Epi 6h | 723.964 | 131.089 | 0.629196 | 0.170383 |
| SA 6h | 681.035 | 93.912 | 0.684568 | 0.0685457 |
| Me-JA 6h | 1087.6 | 180.439 | 1.07619 | 0.0888902 |
Source Transcript PGSC0003DMT400075850 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G26310.1 | +3 | 5e-40 | 127 | 102/374 (27%) | UDP-Glycosyltransferase superfamily protein | chr5:9234739-9236184 FORWARD LENGTH=481 |
