Probe CUST_29447_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29447_PI426222305 | JHI_St_60k_v1 | DMT400080576 | ATGTTCAGAGAGTTGAGATAAAGCAATTCTCACTGGGGGATGAGCCTCTCTCTGTTAGGA |
All Microarray Probes Designed to Gene DMG400031377
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29447_PI426222305 | JHI_St_60k_v1 | DMT400080576 | ATGTTCAGAGAGTTGAGATAAAGCAATTCTCACTGGGGGATGAGCCTCTCTCTGTTAGGA |
Microarray Signals from CUST_29447_PI426222305
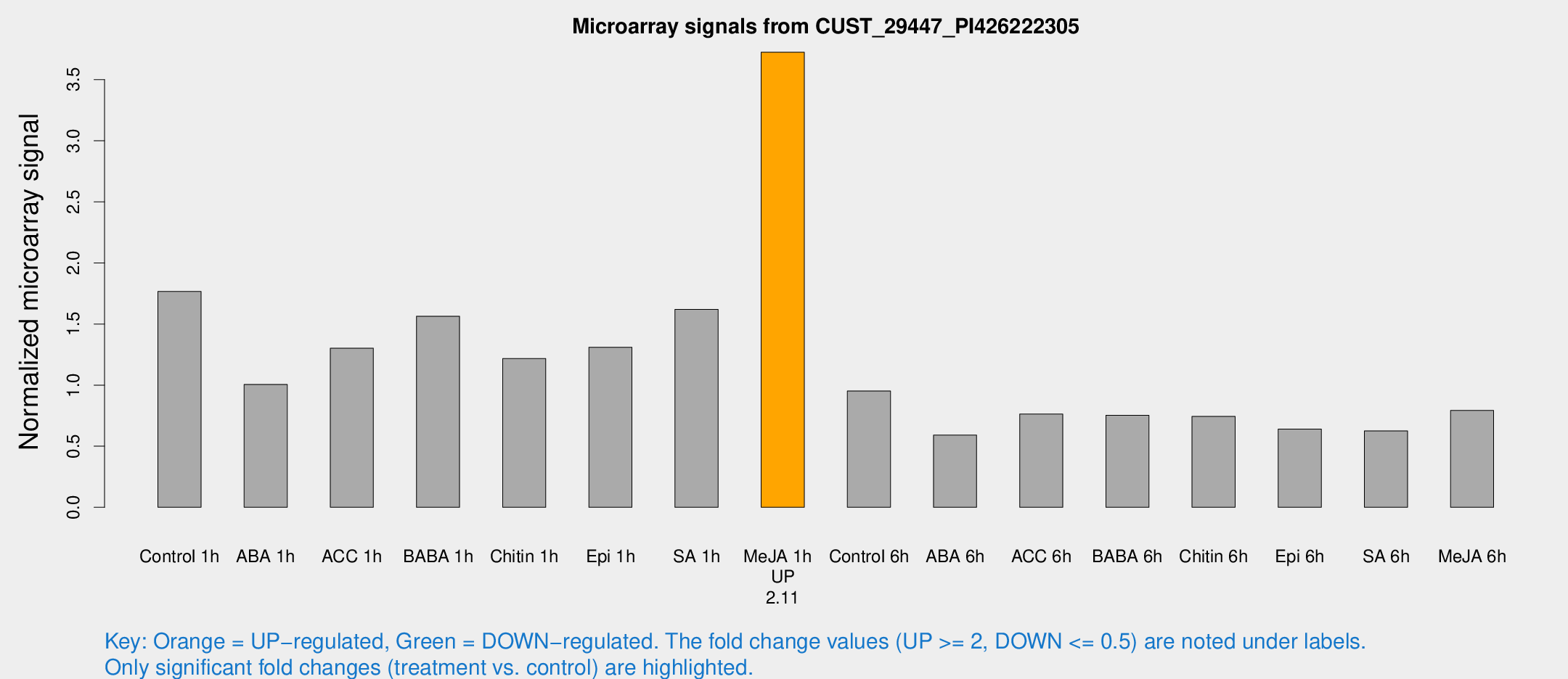
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 717.388 | 62.6437 | 1.7662 | 0.10238 |
| ABA 1h | 362.191 | 35.4573 | 1.00549 | 0.15994 |
| ACC 1h | 571.259 | 134.146 | 1.302 | 0.244618 |
| BABA 1h | 620.504 | 88.5302 | 1.56385 | 0.109719 |
| Chitin 1h | 459.858 | 95.5673 | 1.21769 | 0.297415 |
| Epi 1h | 461.16 | 41.5137 | 1.31011 | 0.159654 |
| SA 1h | 747.728 | 259.197 | 1.61938 | 0.577534 |
| Me-JA 1h | 1240.6 | 139.147 | 3.72444 | 0.235521 |
| Control 6h | 395.423 | 60.9132 | 0.952278 | 0.0712182 |
| ABA 6h | 268.751 | 58.8283 | 0.59093 | 0.126698 |
| ACC 6h | 353.334 | 20.9134 | 0.762692 | 0.10445 |
| BABA 6h | 354.499 | 68.2116 | 0.753308 | 0.130809 |
| Chitin 6h | 322.926 | 33.0656 | 0.743999 | 0.0878688 |
| Epi 6h | 301.872 | 61.2368 | 0.639498 | 0.129458 |
| SA 6h | 289.922 | 108.462 | 0.62469 | 0.387941 |
| Me-JA 6h | 325.432 | 49.4738 | 0.792756 | 0.0567486 |
Source Transcript PGSC0003DMT400080576 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G19830.1 | +3 | 0.0 | 726 | 406/631 (64%) | Calcium-dependent lipid-binding (CaLB domain) family protein | chr3:6886338-6889974 REVERSE LENGTH=693 |
