Probe CUST_29415_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29415_PI426222305 | JHI_St_60k_v1 | DMT400075696 | TAAAGAGGTCTGACCGACTATATCATGTACCTTATATTTCAGGCATGGAAGTTATGGAAA |
All Microarray Probes Designed to Gene DMG401029444
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29461_PI426222305 | JHI_St_60k_v1 | DMT400075699 | GCACCATCACAACCTGCTTTTACTGAAAGACATGATTGTATCTTCAAAATGTGCAATGTA |
| CUST_29494_PI426222305 | JHI_St_60k_v1 | DMT400075694 | AGAGTAGTTGGTACTTATGGATACATGGCCCCTGAATATGCAATGAAAGGAAGATTCTCT |
| CUST_29468_PI426222305 | JHI_St_60k_v1 | DMT400075700 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
| CUST_29464_PI426222305 | JHI_St_60k_v1 | DMT400075698 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
| CUST_29383_PI426222305 | JHI_St_60k_v1 | DMT400075701 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
| CUST_29415_PI426222305 | JHI_St_60k_v1 | DMT400075696 | TAAAGAGGTCTGACCGACTATATCATGTACCTTATATTTCAGGCATGGAAGTTATGGAAA |
| CUST_29484_PI426222305 | JHI_St_60k_v1 | DMT400075702 | CCATTCGTCTCAATTTGTATGATACCTTTTGTTTCAAAATGTTTGACAATATTGGGTCCG |
Microarray Signals from CUST_29415_PI426222305
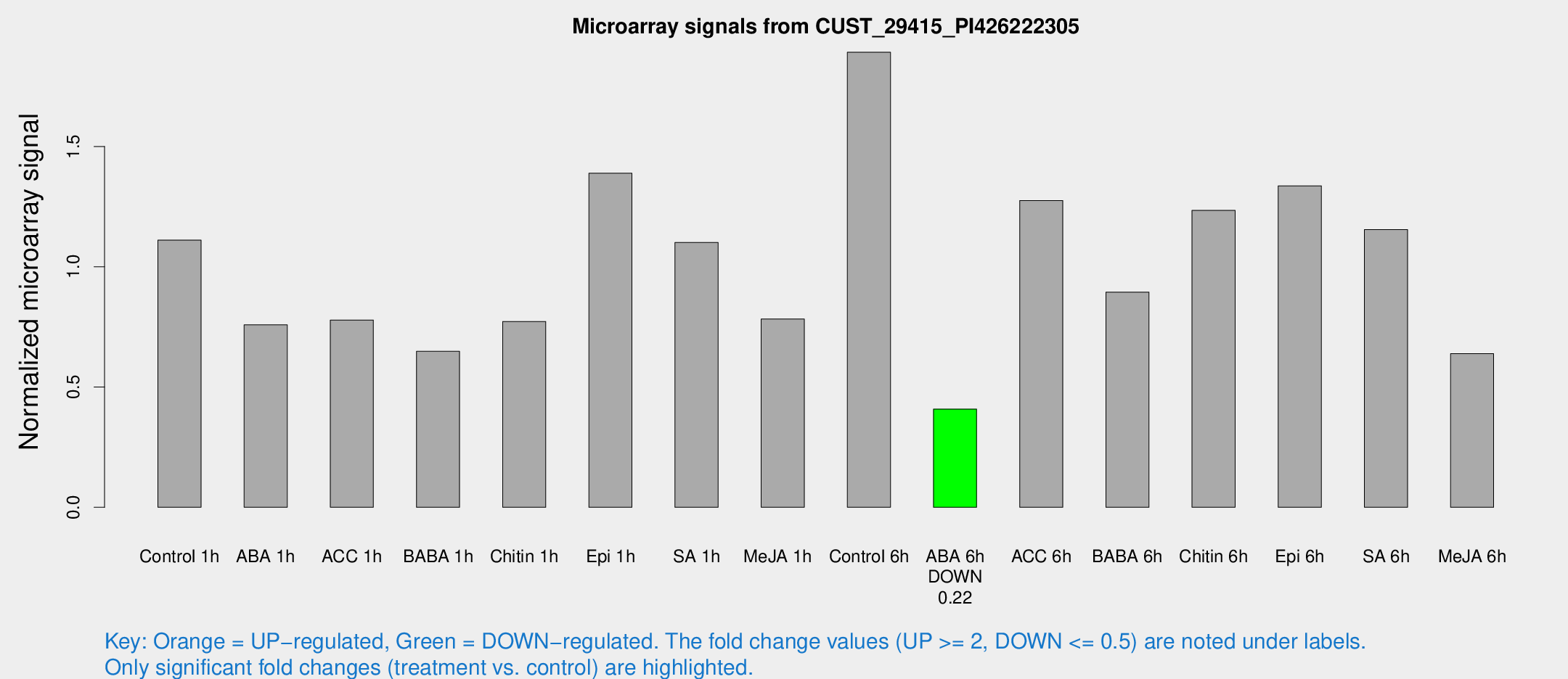
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 21.2952 | 4.90699 | 1.11064 | 0.225164 |
| ABA 1h | 12.9936 | 3.61742 | 0.75849 | 0.234638 |
| ACC 1h | 15.0749 | 4.63122 | 0.778278 | 0.277228 |
| BABA 1h | 13.9563 | 6.45595 | 0.648338 | 0.270875 |
| Chitin 1h | 13.381 | 3.73422 | 0.77248 | 0.236381 |
| Epi 1h | 22.2947 | 3.80127 | 1.38853 | 0.247 |
| SA 1h | 21.1091 | 3.7765 | 1.10118 | 0.208354 |
| Me-JA 1h | 12.7538 | 3.84594 | 0.782972 | 0.289798 |
| Control 6h | 37.3675 | 10.5223 | 1.89172 | 0.382955 |
| ABA 6h | 7.99825 | 3.90205 | 0.408408 | 0.202787 |
| ACC 6h | 28.1825 | 6.2762 | 1.27523 | 0.233441 |
| BABA 6h | 19.2958 | 4.34484 | 0.894713 | 0.234856 |
| Chitin 6h | 24.3682 | 4.38123 | 1.23399 | 0.228636 |
| Epi 6h | 33.8014 | 12.7875 | 1.33644 | 0.928976 |
| SA 6h | 21.2257 | 4.21721 | 1.1541 | 0.241676 |
| Me-JA 6h | 14.364 | 6.76755 | 0.638514 | 0.264686 |
Source Transcript PGSC0003DMT400075696 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G11330.2 | +1 | 0.0 | 680 | 356/728 (49%) | S-locus lectin protein kinase family protein | chr1:3810372-3813416 FORWARD LENGTH=842 |
