Probe CUST_29161_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29161_PI426222305 | JHI_St_60k_v1 | DMT400020669 | CCAATTTGCACCGTGATAGTAAGTGGCTTATGTTGAAGATAACTATCTTCTAGTAAAAGG |
All Microarray Probes Designed to Gene DMG400008011
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29161_PI426222305 | JHI_St_60k_v1 | DMT400020669 | CCAATTTGCACCGTGATAGTAAGTGGCTTATGTTGAAGATAACTATCTTCTAGTAAAAGG |
| CUST_29022_PI426222305 | JHI_St_60k_v1 | DMT400020670 | ATTTGCACCGTGATAGTAAGTGGCTTATGTTGAAGATAACTATCTTCTAGTAAAAGGGCA |
| CUST_29143_PI426222305 | JHI_St_60k_v1 | DMT400020671 | CCCTATAACCTGCATTTTAATCTCACTATGTACTTCATCTAGTCAGGCCATCCATGTTAT |
Microarray Signals from CUST_29161_PI426222305
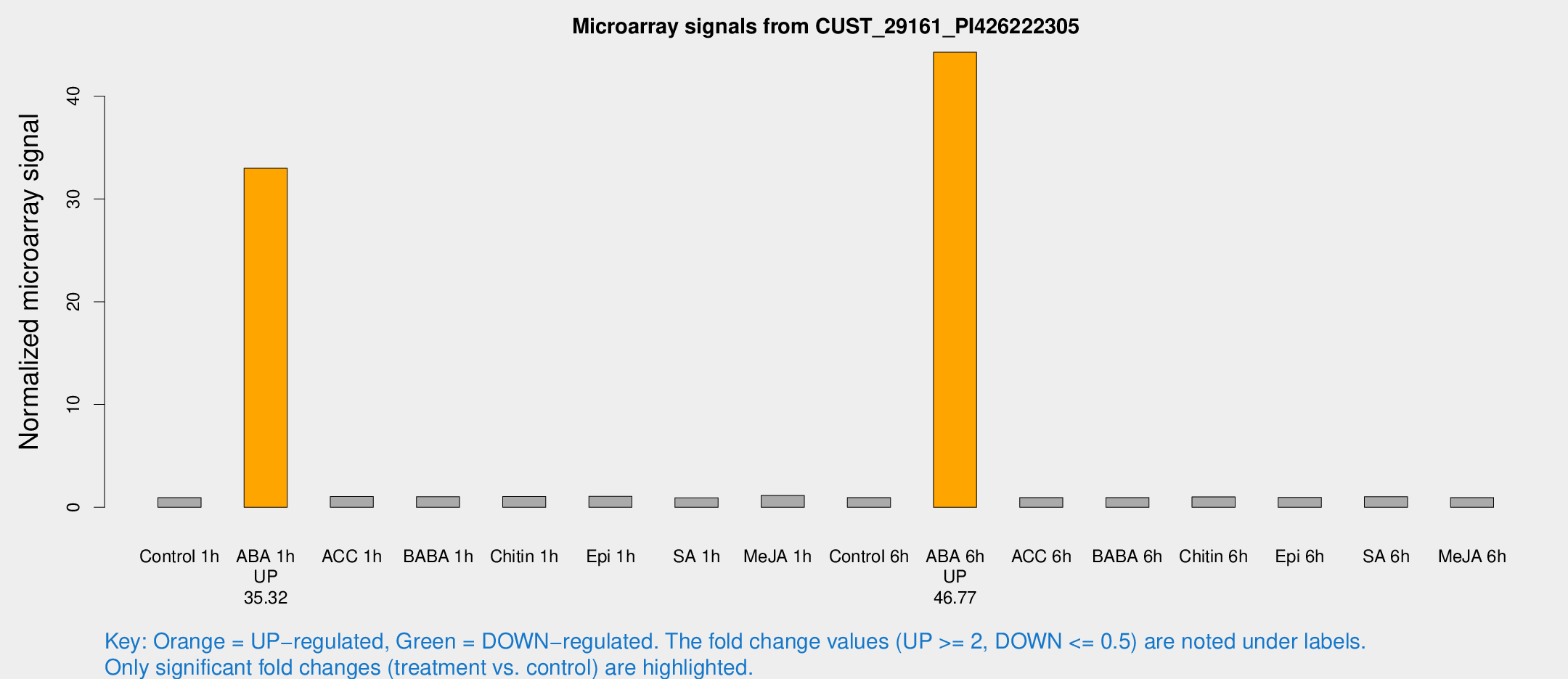
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.26071 | 3.05032 | 0.933865 | 0.536245 |
| ABA 1h | 173.506 | 37.3688 | 32.9843 | 6.715 |
| ACC 1h | 6.18887 | 3.62868 | 1.04854 | 0.606309 |
| BABA 1h | 5.5889 | 3.24468 | 1.02003 | 0.591066 |
| Chitin 1h | 5.20861 | 3.03453 | 1.04974 | 0.588342 |
| Epi 1h | 5.14382 | 2.98768 | 1.06961 | 0.606159 |
| SA 1h | 5.3316 | 3.07899 | 0.914585 | 0.527955 |
| Me-JA 1h | 5.351 | 3.11381 | 1.15359 | 0.66586 |
| Control 6h | 5.38618 | 3.1234 | 0.94655 | 0.548538 |
| ABA 6h | 285.564 | 65.6154 | 44.2664 | 10.5649 |
| ACC 6h | 6.28363 | 3.72776 | 0.943743 | 0.546573 |
| BABA 6h | 6.01558 | 3.48979 | 0.94556 | 0.547556 |
| Chitin 6h | 6.12274 | 3.46983 | 1.01305 | 0.574391 |
| Epi 6h | 6.21485 | 3.64348 | 0.961639 | 0.558063 |
| SA 6h | 5.74541 | 3.32787 | 1.02356 | 0.592724 |
| Me-JA 6h | 5.3517 | 3.10366 | 0.947118 | 0.54917 |
Source Transcript PGSC0003DMT400020669 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
