Probe CUST_29052_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29052_PI426222305 | JHI_St_60k_v1 | DMT400020700 | GCACAACGGAAAAAGGTCAGAAATGCTCTTAATTTTTCATTAAACACGCGACCTTTATTT |
All Microarray Probes Designed to Gene DMG400008022
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29052_PI426222305 | JHI_St_60k_v1 | DMT400020700 | GCACAACGGAAAAAGGTCAGAAATGCTCTTAATTTTTCATTAAACACGCGACCTTTATTT |
Microarray Signals from CUST_29052_PI426222305
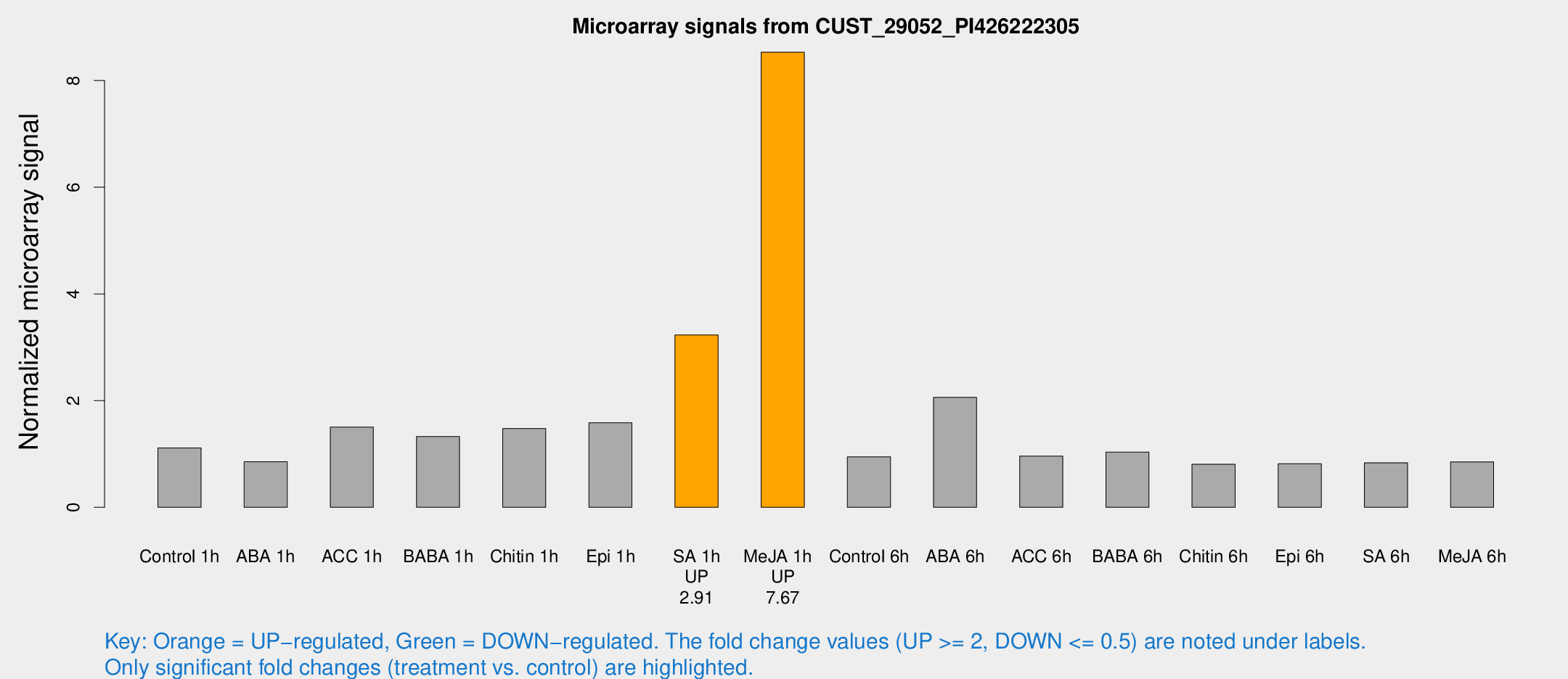
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.98265 | 3.75204 | 1.11179 | 0.462018 |
| ABA 1h | 6.47827 | 3.67271 | 0.854544 | 0.484674 |
| ACC 1h | 13.9792 | 4.09984 | 1.50361 | 0.504563 |
| BABA 1h | 11.556 | 4.06906 | 1.32649 | 0.52917 |
| Chitin 1h | 12.0439 | 3.89827 | 1.47756 | 0.559198 |
| Epi 1h | 12.1793 | 3.74136 | 1.5842 | 0.529649 |
| SA 1h | 30.6938 | 8.801 | 3.2304 | 0.886516 |
| Me-JA 1h | 60.206 | 6.30788 | 8.53026 | 0.97333 |
| Control 6h | 8.33885 | 3.81978 | 0.945203 | 0.466165 |
| ABA 6h | 48.8459 | 41.7611 | 2.06239 | 4.16189 |
| ACC 6h | 9.71254 | 4.77833 | 0.959137 | 0.482516 |
| BABA 6h | 10.4803 | 4.3791 | 1.03286 | 0.493947 |
| Chitin 6h | 7.36161 | 4.26652 | 0.807996 | 0.468022 |
| Epi 6h | 7.94015 | 4.64148 | 0.817037 | 0.474226 |
| SA 6h | 7.05525 | 4.08583 | 0.833776 | 0.482824 |
| Me-JA 6h | 7.30809 | 3.78928 | 0.851991 | 0.449766 |
Source Transcript PGSC0003DMT400020700 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G19184.1 | +3 | 1e-57 | 192 | 110/220 (50%) | AP2/B3-like transcriptional factor family protein | chr3:6637555-6639035 FORWARD LENGTH=277 |
