Probe CUST_2895_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2895_PI426222305 | JHI_St_60k_v1 | DMT400000572 | CAAAAGGAAAACTTTCAGATGACCAGGCCTTACATAGGATTTGGTTCCACGGACCTTTGA |
All Microarray Probes Designed to Gene DMG400000200
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2895_PI426222305 | JHI_St_60k_v1 | DMT400000572 | CAAAAGGAAAACTTTCAGATGACCAGGCCTTACATAGGATTTGGTTCCACGGACCTTTGA |
Microarray Signals from CUST_2895_PI426222305
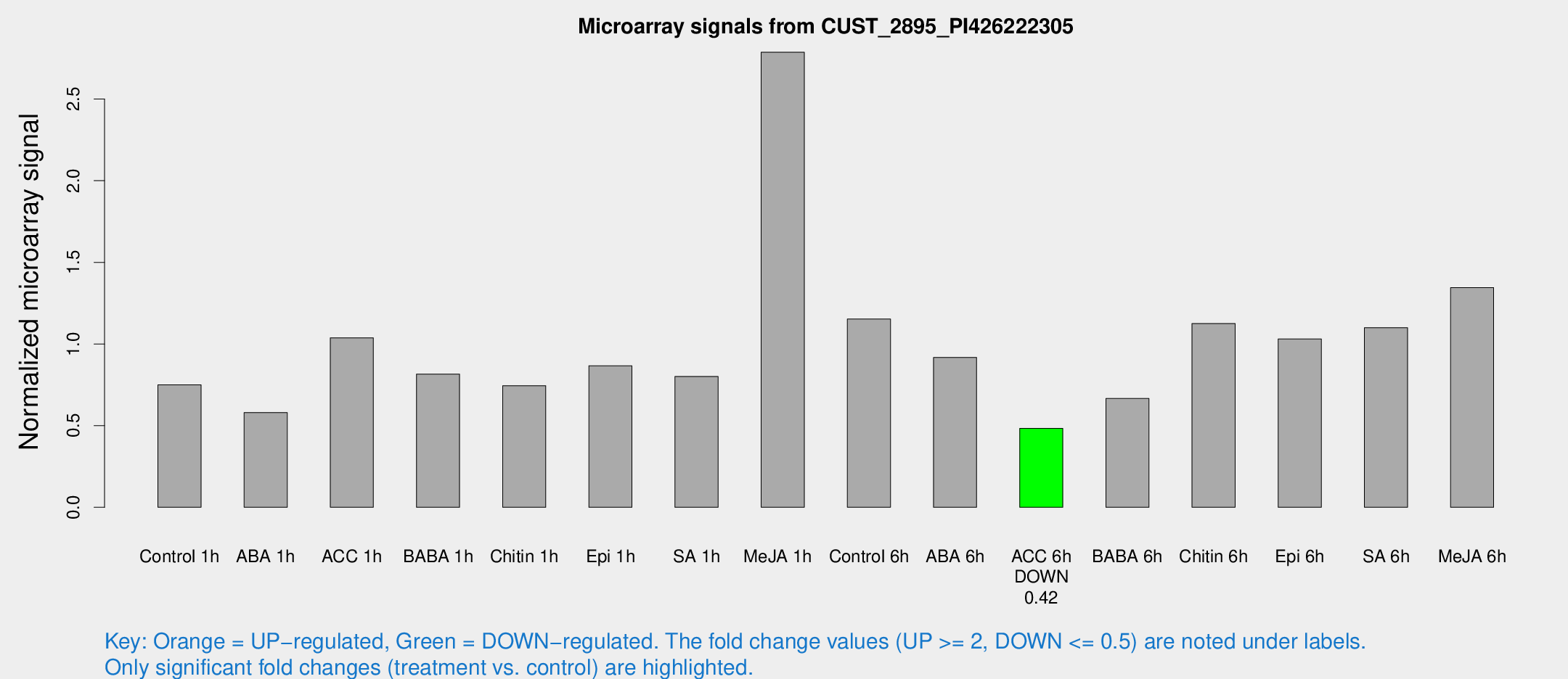
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 18.122 | 4.21202 | 0.749935 | 0.176821 |
| ABA 1h | 14.1779 | 6.13569 | 0.580397 | 0.229606 |
| ACC 1h | 25.2091 | 4.3514 | 1.03818 | 0.208493 |
| BABA 1h | 19.7616 | 5.1619 | 0.815897 | 0.21311 |
| Chitin 1h | 16.211 | 3.66379 | 0.744729 | 0.247361 |
| Epi 1h | 17.9277 | 3.67564 | 0.866466 | 0.193078 |
| SA 1h | 25.1153 | 11.165 | 0.801256 | 0.555169 |
| Me-JA 1h | 60.1099 | 23.4682 | 2.78683 | 0.842236 |
| Control 6h | 29.0033 | 7.6683 | 1.15316 | 0.274077 |
| ABA 6h | 26.7801 | 11.0594 | 0.917855 | 0.381372 |
| ACC 6h | 12.8018 | 4.54939 | 0.483553 | 0.169633 |
| BABA 6h | 17.1377 | 4.17757 | 0.666449 | 0.164165 |
| Chitin 6h | 28.0448 | 4.52632 | 1.12506 | 0.1894 |
| Epi 6h | 26.8543 | 4.66742 | 1.03099 | 0.179564 |
| SA 6h | 27.0777 | 6.74841 | 1.10016 | 0.223088 |
| Me-JA 6h | 31.1102 | 4.11654 | 1.34535 | 0.181195 |
Source Transcript PGSC0003DMT400000572 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G01710.1 | +1 | 3e-59 | 192 | 101/174 (58%) | Acyl-CoA thioesterase family protein | chr1:262950-266029 FORWARD LENGTH=427 |
