Probe CUST_28897_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28897_PI426222305 | JHI_St_60k_v1 | DMT400033336 | CATGCTTGTGGCATCAATAGATACTTTTTAAATTTAAATTCAGGACATGCTTGTGGCATC |
All Microarray Probes Designed to Gene DMG402012803
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28897_PI426222305 | JHI_St_60k_v1 | DMT400033336 | CATGCTTGTGGCATCAATAGATACTTTTTAAATTTAAATTCAGGACATGCTTGTGGCATC |
| CUST_28888_PI426222305 | JHI_St_60k_v1 | DMT400033335 | GGCTGTTCCAACTTATAGGCTACAACACAACTGATTTTGGGAAATTGGTTTTGATATATA |
Microarray Signals from CUST_28897_PI426222305
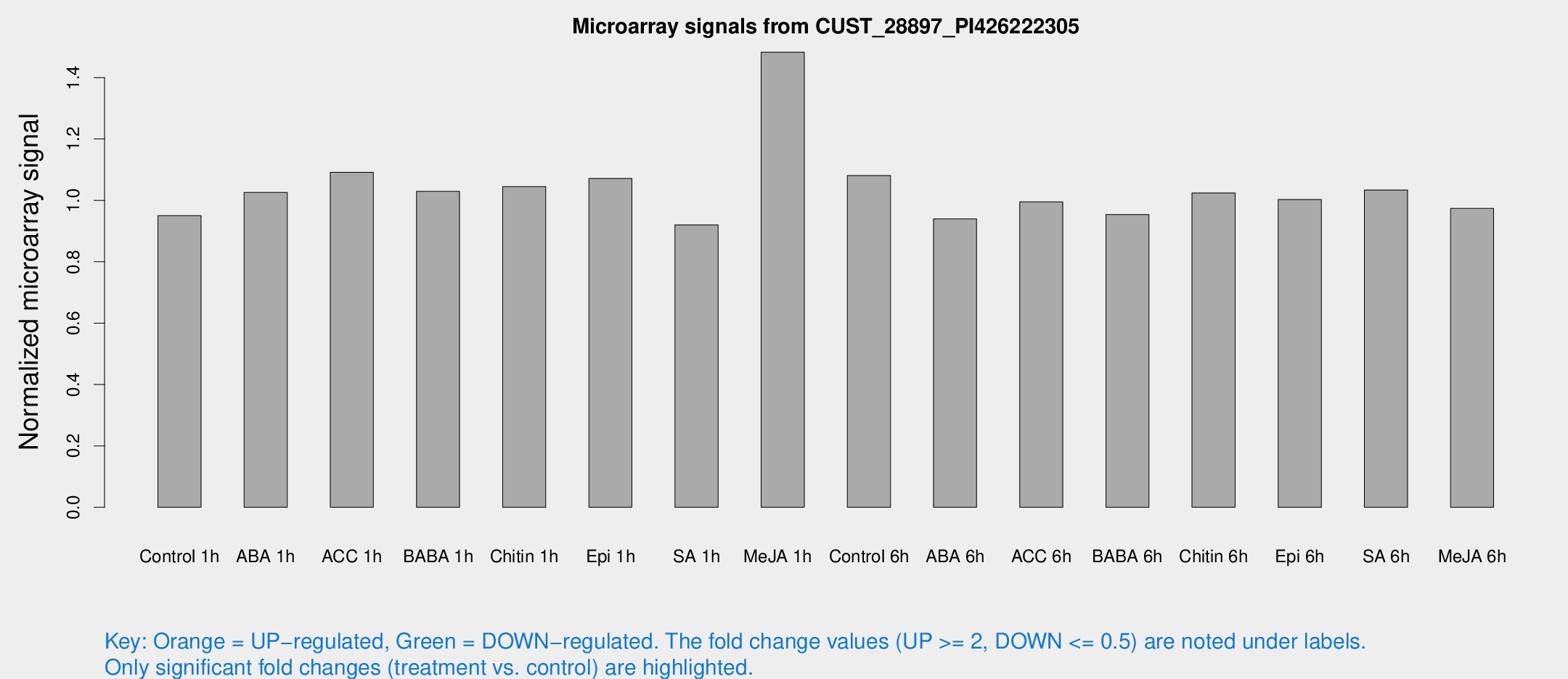
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.83694 | 3.38509 | 0.950529 | 0.550532 |
| ABA 1h | 5.57702 | 3.24155 | 1.026 | 0.595069 |
| ACC 1h | 7.02495 | 4.16361 | 1.0915 | 0.632584 |
| BABA 1h | 6.08809 | 3.53161 | 1.0294 | 0.596792 |
| Chitin 1h | 5.77596 | 3.35226 | 1.04501 | 0.60516 |
| Epi 1h | 5.69585 | 3.30331 | 1.07141 | 0.62042 |
| SA 1h | 5.79808 | 3.35869 | 0.920012 | 0.532764 |
| Me-JA 1h | 7.73493 | 3.47567 | 1.48273 | 0.728521 |
| Control 6h | 6.71691 | 3.45645 | 1.08073 | 0.569277 |
| ABA 6h | 6.13414 | 3.55708 | 0.939451 | 0.543998 |
| ACC 6h | 7.18997 | 4.28129 | 0.995302 | 0.576595 |
| BABA 6h | 6.5641 | 3.80945 | 0.954008 | 0.552448 |
| Chitin 6h | 6.68686 | 3.8743 | 1.02392 | 0.593049 |
| Epi 6h | 6.98029 | 4.06906 | 1.00278 | 0.580666 |
| SA 6h | 6.27528 | 3.63631 | 1.03381 | 0.598695 |
| Me-JA 6h | 5.95564 | 3.45633 | 0.973801 | 0.564342 |
Source Transcript PGSC0003DMT400033336 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G36220.1 | +1 | 6e-100 | 316 | 176/467 (38%) | ferulic acid 5-hydroxylase 1 | chr4:17137584-17139619 REVERSE LENGTH=520 |
