Probe CUST_28818_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28818_PI426222305 | JHI_St_60k_v1 | DMT400083032 | AATGAATCCTCTAAAGTGGCTCAGGATGAAGGTTATGCCTCTGCAGATGACGCTGTTCAA |
All Microarray Probes Designed to Gene DMG400033030
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28818_PI426222305 | JHI_St_60k_v1 | DMT400083032 | AATGAATCCTCTAAAGTGGCTCAGGATGAAGGTTATGCCTCTGCAGATGACGCTGTTCAA |
Microarray Signals from CUST_28818_PI426222305
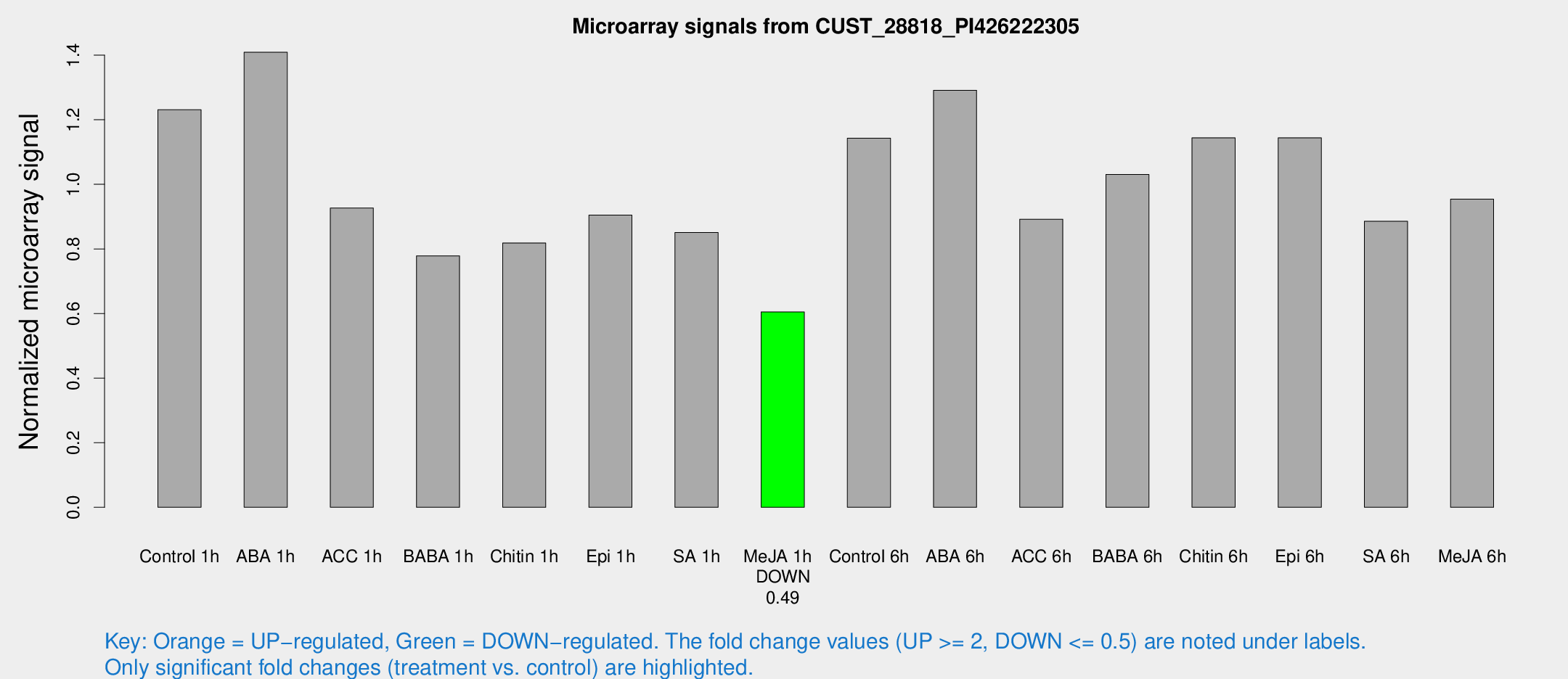
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5790.04 | 481.564 | 1.23092 | 0.0710713 |
| ABA 1h | 5936.81 | 804.169 | 1.40902 | 0.103294 |
| ACC 1h | 4702.14 | 1003.4 | 0.926578 | 0.159257 |
| BABA 1h | 3678.08 | 810.987 | 0.778485 | 0.113541 |
| Chitin 1h | 3500.2 | 484.628 | 0.818357 | 0.0612882 |
| Epi 1h | 3702.05 | 389.263 | 0.90456 | 0.0911054 |
| SA 1h | 4190.8 | 693.566 | 0.850998 | 0.0866591 |
| Me-JA 1h | 2317.69 | 156.156 | 0.604816 | 0.0349327 |
| Control 6h | 5649.08 | 1247.7 | 1.143 | 0.187836 |
| ABA 6h | 6573.23 | 968.316 | 1.29116 | 0.16071 |
| ACC 6h | 4954.02 | 907.393 | 0.892137 | 0.0824277 |
| BABA 6h | 5500.7 | 810.345 | 1.03065 | 0.138844 |
| Chitin 6h | 5725.35 | 440.322 | 1.14415 | 0.0830902 |
| Epi 6h | 6059.68 | 443.932 | 1.14407 | 0.0783065 |
| SA 6h | 4139.28 | 451.913 | 0.885541 | 0.0883261 |
| Me-JA 6h | 4777.23 | 1343.5 | 0.953992 | 0.18546 |
Source Transcript PGSC0003DMT400083032 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G19000.1 | +2 | 1e-52 | 181 | 149/310 (48%) | Homeodomain-like superfamily protein | chr1:6561335-6562684 REVERSE LENGTH=285 |
