Probe CUST_28539_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28539_PI426222305 | JHI_St_60k_v1 | DMT400055531 | GTCTCTCCCAATATGAGAATCAGATCTTGTGACATAGAATATATAGATAGTTGTCATGAC |
All Microarray Probes Designed to Gene DMG402021564
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28539_PI426222305 | JHI_St_60k_v1 | DMT400055531 | GTCTCTCCCAATATGAGAATCAGATCTTGTGACATAGAATATATAGATAGTTGTCATGAC |
Microarray Signals from CUST_28539_PI426222305
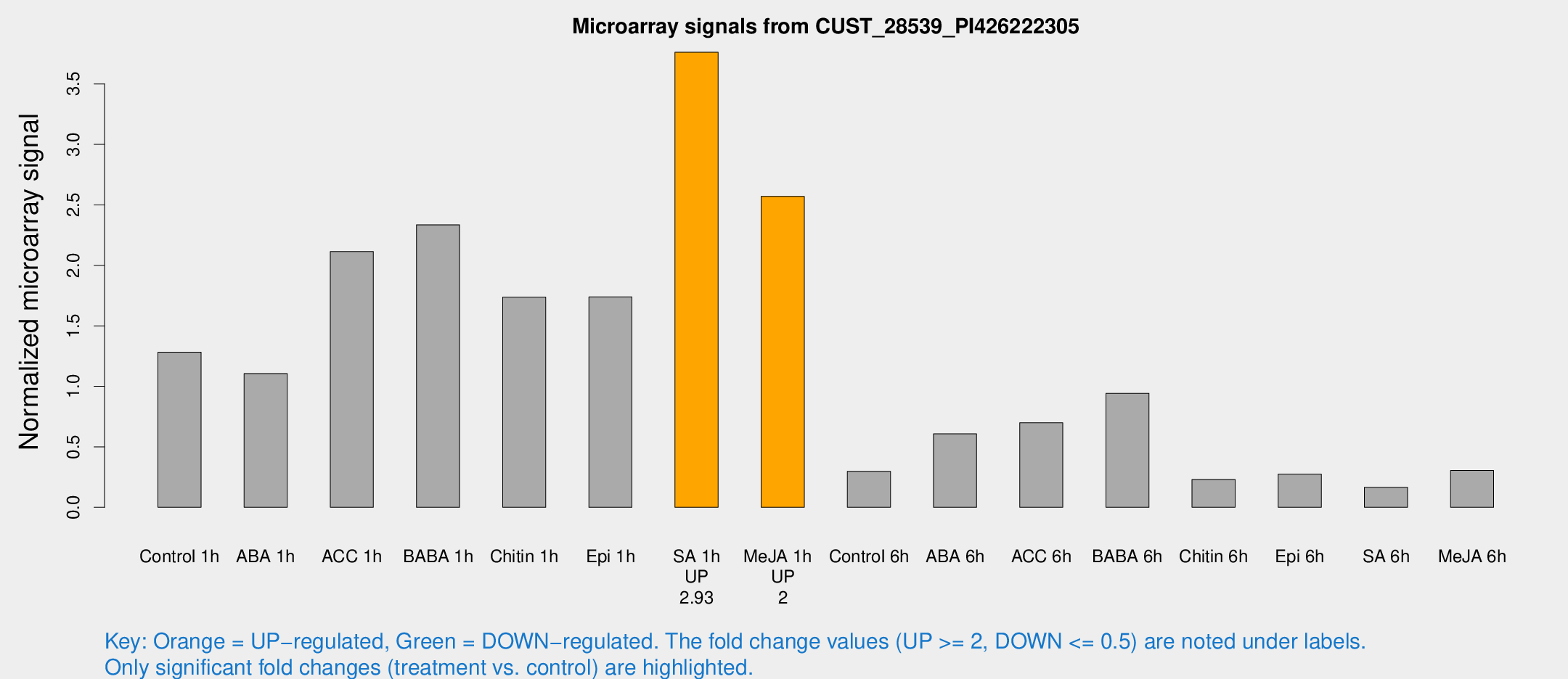
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2335.82 | 457.949 | 1.28223 | 0.215718 |
| ABA 1h | 1721.8 | 108.836 | 1.10566 | 0.0638612 |
| ACC 1h | 4723 | 1730.68 | 2.11385 | 1.08437 |
| BABA 1h | 4016.53 | 505.065 | 2.33547 | 0.134852 |
| Chitin 1h | 2792.12 | 381.408 | 1.73782 | 0.205534 |
| Epi 1h | 2654.7 | 197.88 | 1.73898 | 0.122945 |
| SA 1h | 7168.23 | 1712.58 | 3.76196 | 0.842359 |
| Me-JA 1h | 3736.86 | 477.906 | 2.57043 | 0.18082 |
| Control 6h | 625.874 | 270.476 | 0.297614 | 0.1109 |
| ABA 6h | 1214.43 | 320.483 | 0.607003 | 0.132263 |
| ACC 6h | 1432.86 | 195.78 | 0.698447 | 0.143227 |
| BABA 6h | 1929.08 | 397.456 | 0.942693 | 0.183418 |
| Chitin 6h | 432.891 | 33.346 | 0.230448 | 0.0187914 |
| Epi 6h | 552 | 64.2738 | 0.275113 | 0.0208603 |
| SA 6h | 288.728 | 29.6436 | 0.164755 | 0.0126411 |
| Me-JA 6h | 592.339 | 199.198 | 0.305225 | 0.0766709 |
Source Transcript PGSC0003DMT400055531 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37040.1 | +1 | 0.0 | 1167 | 571/699 (82%) | PHE ammonia lyase 1 | chr2:15557602-15560237 REVERSE LENGTH=725 |
