Probe CUST_28467_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28467_PI426222305 | JHI_St_60k_v1 | DMT400074600 | TTCCTTTGATTGTTGATTTGCAGGTGATGAATAGTTTCGTTCATTCCTTACCTATGTTGC |
All Microarray Probes Designed to Gene DMG400029005
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28467_PI426222305 | JHI_St_60k_v1 | DMT400074600 | TTCCTTTGATTGTTGATTTGCAGGTGATGAATAGTTTCGTTCATTCCTTACCTATGTTGC |
Microarray Signals from CUST_28467_PI426222305
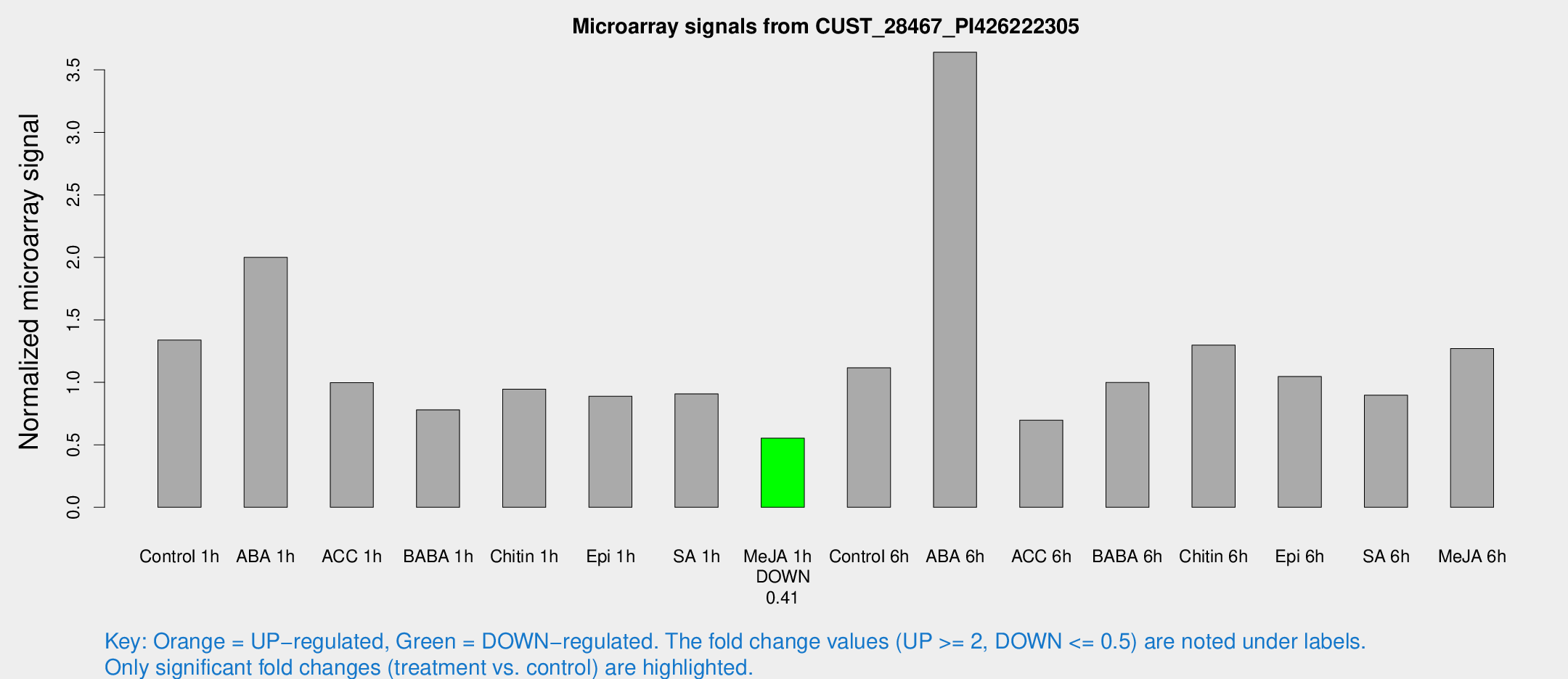
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 148.561 | 12.3046 | 1.33841 | 0.0842019 |
| ABA 1h | 197.596 | 21.5655 | 1.99991 | 0.121351 |
| ACC 1h | 113.414 | 7.72658 | 0.997223 | 0.0681103 |
| BABA 1h | 87.4783 | 18.2801 | 0.780018 | 0.108221 |
| Chitin 1h | 99.321 | 22.0124 | 0.945722 | 0.168424 |
| Epi 1h | 85.5965 | 7.77908 | 0.889057 | 0.0700063 |
| SA 1h | 106.313 | 18.0194 | 0.907775 | 0.130163 |
| Me-JA 1h | 51.8523 | 10.289 | 0.553304 | 0.064066 |
| Control 6h | 149.31 | 57.158 | 1.11645 | 0.479916 |
| ABA 6h | 450.996 | 96.8617 | 3.64146 | 0.726417 |
| ACC 6h | 92.3848 | 18.1448 | 0.697282 | 0.116343 |
| BABA 6h | 129.133 | 29.0736 | 0.998883 | 0.207256 |
| Chitin 6h | 153.061 | 10.2746 | 1.29806 | 0.108913 |
| Epi 6h | 139.479 | 38.7545 | 1.04627 | 0.280025 |
| SA 6h | 105.826 | 31.0128 | 0.897485 | 0.231558 |
| Me-JA 6h | 154.162 | 42.652 | 1.27075 | 0.338003 |
Source Transcript PGSC0003DMT400074600 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G17230.3 | +1 | 2e-68 | 219 | 110/146 (75%) | PHYTOENE SYNTHASE | chr5:5659839-5662087 REVERSE LENGTH=437 |
