Probe CUST_28361_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28361_PI426222305 | JHI_St_60k_v1 | DMT400044378 | GCTGCGTTATGGATATGTAGTATACTTGGATAATCATGTGTTATGTGCCTTCTTTATAAC |
All Microarray Probes Designed to Gene DMG400017230
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28361_PI426222305 | JHI_St_60k_v1 | DMT400044378 | GCTGCGTTATGGATATGTAGTATACTTGGATAATCATGTGTTATGTGCCTTCTTTATAAC |
Microarray Signals from CUST_28361_PI426222305
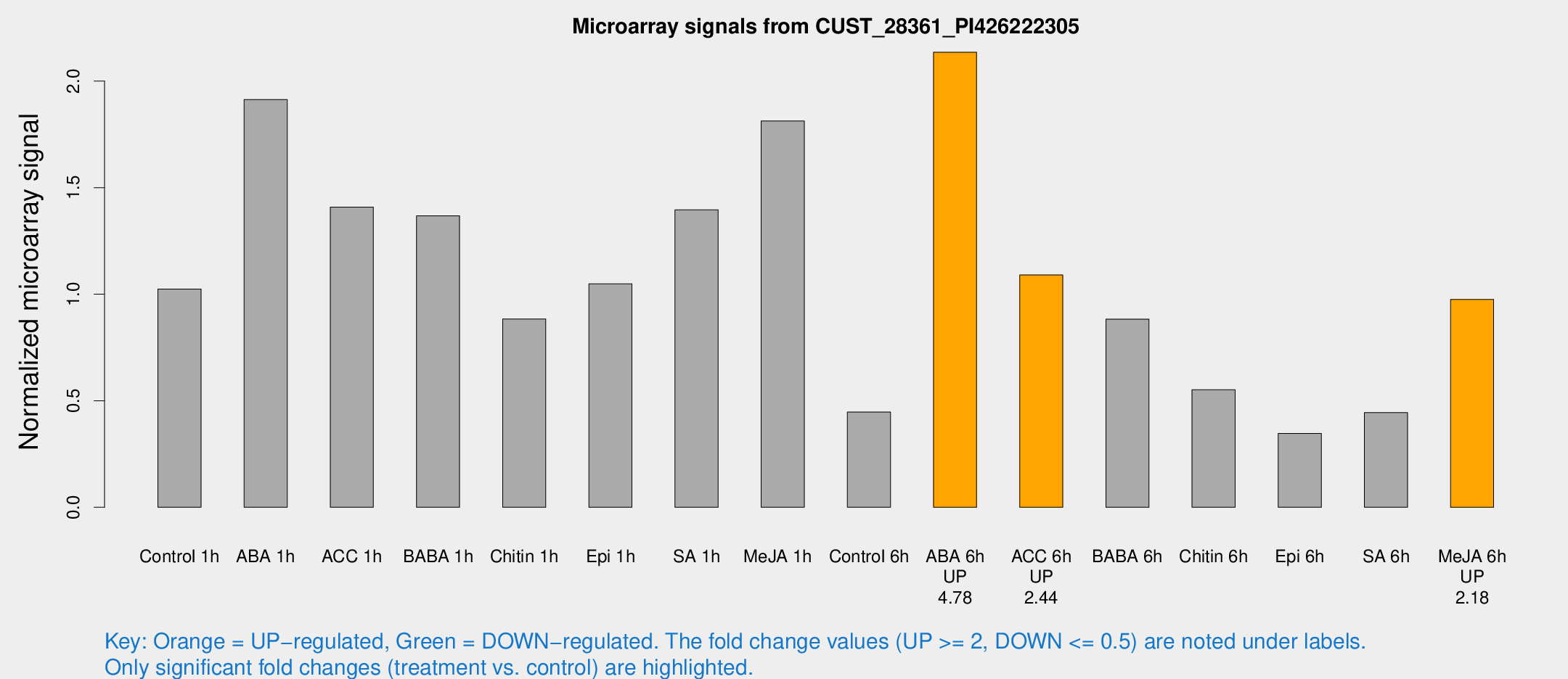
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 373.512 | 87.5303 | 1.02418 | 0.166881 |
| ABA 1h | 668.298 | 201.878 | 1.91363 | 0.705907 |
| ACC 1h | 588.259 | 213.836 | 1.40879 | 0.541313 |
| BABA 1h | 483.089 | 102.377 | 1.36812 | 0.192395 |
| Chitin 1h | 291.512 | 71.7343 | 0.884038 | 0.157417 |
| Epi 1h | 320.092 | 40.4509 | 1.04869 | 0.132902 |
| SA 1h | 503.636 | 53.5823 | 1.39586 | 0.0963204 |
| Me-JA 1h | 547.69 | 137.329 | 1.81332 | 0.310794 |
| Control 6h | 161.08 | 29.6362 | 0.446872 | 0.0460186 |
| ABA 6h | 818.348 | 159.242 | 2.13554 | 0.303848 |
| ACC 6h | 436.044 | 29.3644 | 1.09043 | 0.155337 |
| BABA 6h | 373.992 | 113.136 | 0.883163 | 0.252552 |
| Chitin 6h | 204.077 | 12.3731 | 0.551385 | 0.0338042 |
| Epi 6h | 145.001 | 33.7263 | 0.346301 | 0.0724168 |
| SA 6h | 169.697 | 47.4521 | 0.444121 | 0.107405 |
| Me-JA 6h | 339.144 | 27.0901 | 0.975525 | 0.0571659 |
Source Transcript PGSC0003DMT400044378 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G43400.1 | +3 | 0.0 | 749 | 392/664 (59%) | Uncharacterised conserved protein UCP015417, vWA | chr5:17426182-17428149 REVERSE LENGTH=655 |
