Probe CUST_28007_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28007_PI426222305 | JHI_St_60k_v1 | DMT400082016 | GTGCAAGTGATTAAGCTATCCAAGGATGAAGAGTGAAGATGAAGTGTTTTTAATGCTTAA |
All Microarray Probes Designed to Gene DMG402032203
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28007_PI426222305 | JHI_St_60k_v1 | DMT400082016 | GTGCAAGTGATTAAGCTATCCAAGGATGAAGAGTGAAGATGAAGTGTTTTTAATGCTTAA |
Microarray Signals from CUST_28007_PI426222305
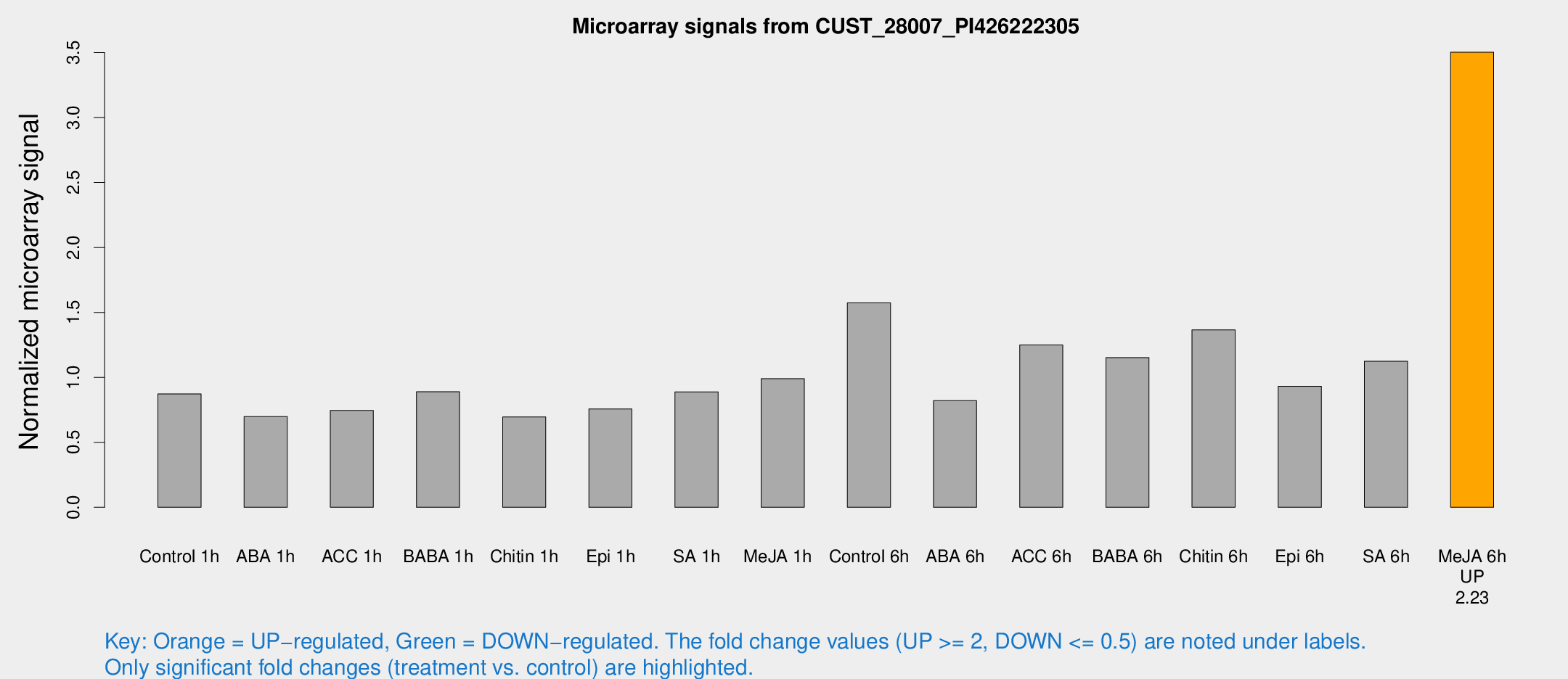
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 24051.1 | 1392.31 | 0.872706 | 0.0503859 |
| ABA 1h | 17755.9 | 3889.63 | 0.698178 | 0.0995618 |
| ACC 1h | 24599 | 7919.56 | 0.745873 | 0.289297 |
| BABA 1h | 24119.4 | 3517.8 | 0.889395 | 0.0540366 |
| Chitin 1h | 17388.5 | 1923.35 | 0.695425 | 0.0401507 |
| Epi 1h | 19043.3 | 4071.82 | 0.756986 | 0.180541 |
| SA 1h | 26135 | 5287.67 | 0.888843 | 0.21523 |
| Me-JA 1h | 22447 | 2172.62 | 0.990749 | 0.0572011 |
| Control 6h | 45819.9 | 10359.5 | 1.57305 | 0.238849 |
| ABA 6h | 24321.6 | 2907.04 | 0.820578 | 0.1296 |
| ACC 6h | 39564.8 | 2442.86 | 1.24991 | 0.121771 |
| BABA 6h | 35479.9 | 2049.46 | 1.15249 | 0.0665391 |
| Chitin 6h | 40120.9 | 2589.98 | 1.36566 | 0.100965 |
| Epi 6h | 29822.4 | 5065.83 | 0.932134 | 0.25107 |
| SA 6h | 31703.2 | 6230.12 | 1.12405 | 0.151604 |
| Me-JA 6h | 98166.8 | 15248.4 | 3.50269 | 0.254416 |
Source Transcript PGSC0003DMT400082016 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
