Probe CUST_27971_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27971_PI426222305 | JHI_St_60k_v1 | DMT400082013 | CACTCTTAATGCTTATTGCGGAATCTATAACGATTGTGATGACGTGCAAGTGATTAAGCT |
All Microarray Probes Designed to Gene DMG401032203
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27943_PI426222305 | JHI_St_60k_v1 | DMT400082012 | AGTGTGCAAGTGATTAAGCTATCCAAGGAGGAGTAAAATTGAAGTGTTTAATTAAGCTGC |
| CUST_27971_PI426222305 | JHI_St_60k_v1 | DMT400082013 | CACTCTTAATGCTTATTGCGGAATCTATAACGATTGTGATGACGTGCAAGTGATTAAGCT |
| CUST_27894_PI426222305 | JHI_St_60k_v1 | DMT400082014 | ACAGTGTGCAAGTGATTAAGCTATCAAAGGATGATGAGTGAAGATGAAGTGTTTTTAATG |
Microarray Signals from CUST_27971_PI426222305
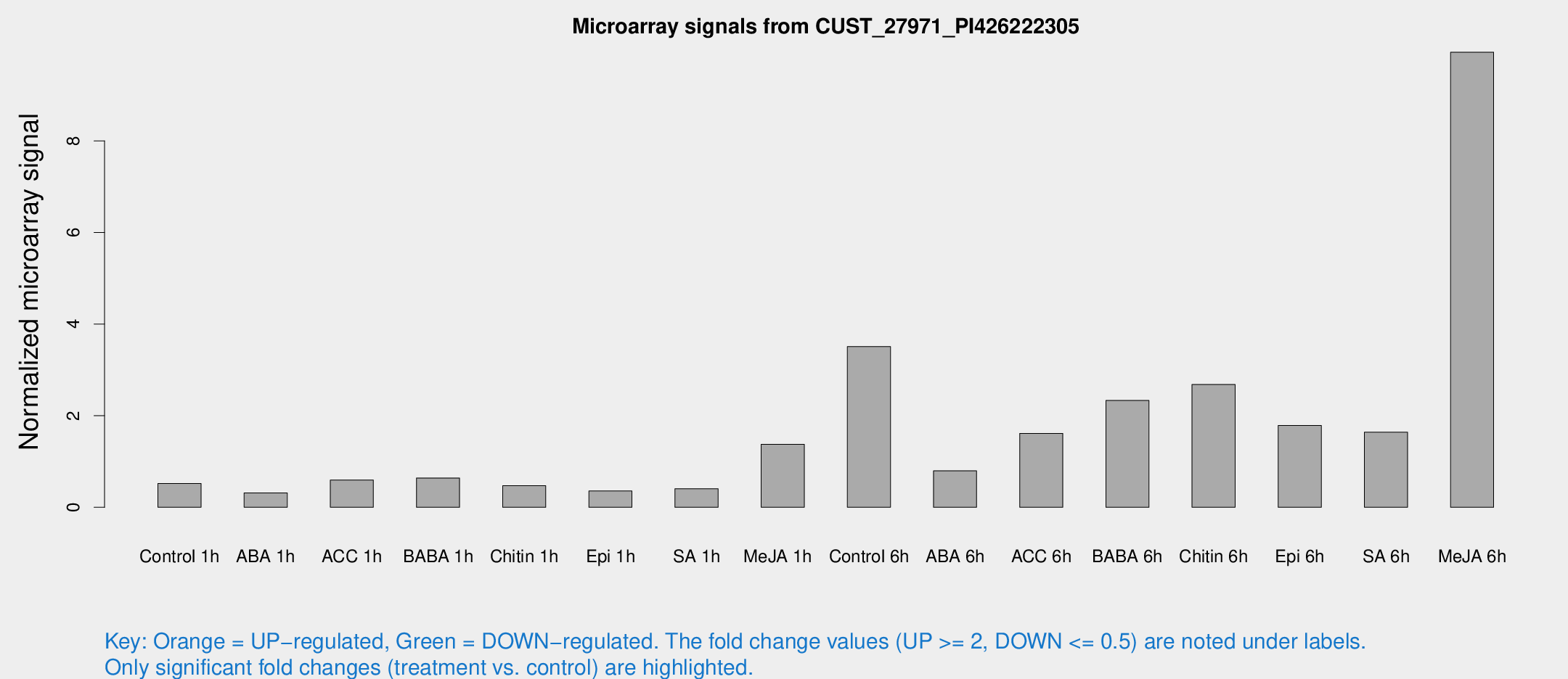
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 29.1984 | 6.42604 | 0.521116 | 0.136736 |
| ABA 1h | 19.1204 | 8.58296 | 0.315389 | 0.191755 |
| ACC 1h | 48.846 | 21.1243 | 0.595537 | 0.585918 |
| BABA 1h | 33.0758 | 4.31454 | 0.640378 | 0.0841634 |
| Chitin 1h | 23.7078 | 5.11809 | 0.472567 | 0.0845893 |
| Epi 1h | 16.9763 | 3.71823 | 0.356041 | 0.0843874 |
| SA 1h | 32.6002 | 16.8422 | 0.405592 | 0.451334 |
| Me-JA 1h | 63.0403 | 14.2633 | 1.3752 | 0.452058 |
| Control 6h | 230.635 | 87.5038 | 3.50973 | 1.57115 |
| ABA 6h | 79.4255 | 56.6512 | 0.796003 | 1.05884 |
| ACC 6h | 114.273 | 36.4892 | 1.61395 | 0.644424 |
| BABA 6h | 151.238 | 44.231 | 2.33135 | 0.592945 |
| Chitin 6h | 173.82 | 64.7362 | 2.68041 | 1.08529 |
| Epi 6h | 139.748 | 53.8392 | 1.78768 | 1.63218 |
| SA 6h | 91.8516 | 21.3765 | 1.63841 | 0.504049 |
| Me-JA 6h | 552.739 | 115.277 | 9.93432 | 1.27759 |
Source Transcript PGSC0003DMT400082013 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
