Probe CUST_27943_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27943_PI426222305 | JHI_St_60k_v1 | DMT400082012 | AGTGTGCAAGTGATTAAGCTATCCAAGGAGGAGTAAAATTGAAGTGTTTAATTAAGCTGC |
All Microarray Probes Designed to Gene DMG401032203
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27971_PI426222305 | JHI_St_60k_v1 | DMT400082013 | CACTCTTAATGCTTATTGCGGAATCTATAACGATTGTGATGACGTGCAAGTGATTAAGCT |
| CUST_27894_PI426222305 | JHI_St_60k_v1 | DMT400082014 | ACAGTGTGCAAGTGATTAAGCTATCAAAGGATGATGAGTGAAGATGAAGTGTTTTTAATG |
| CUST_27943_PI426222305 | JHI_St_60k_v1 | DMT400082012 | AGTGTGCAAGTGATTAAGCTATCCAAGGAGGAGTAAAATTGAAGTGTTTAATTAAGCTGC |
Microarray Signals from CUST_27943_PI426222305
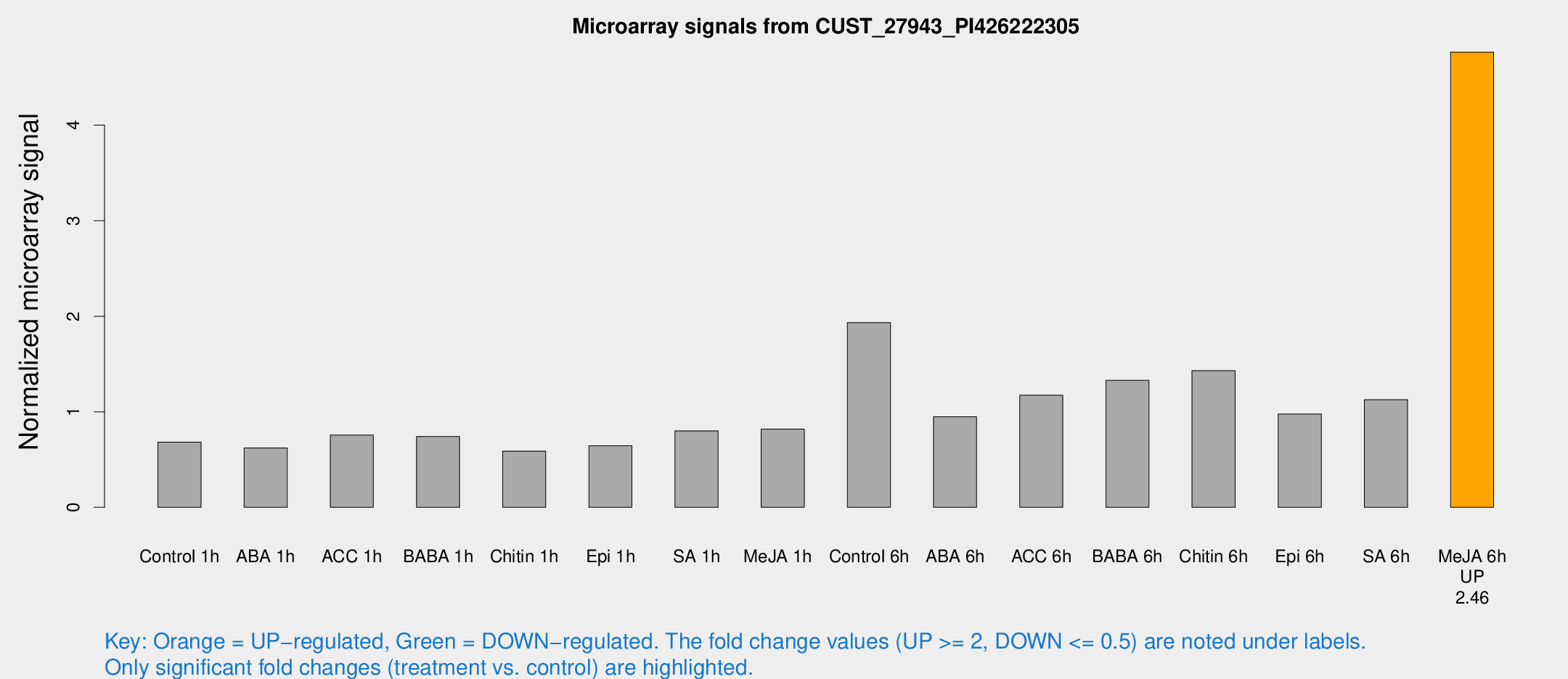
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 882.413 | 160.639 | 0.68237 | 0.083177 |
| ABA 1h | 730.487 | 159.273 | 0.621245 | 0.122187 |
| ACC 1h | 1269.02 | 486.217 | 0.756807 | 0.471033 |
| BABA 1h | 957.088 | 245.463 | 0.7411 | 0.136349 |
| Chitin 1h | 704.819 | 182.856 | 0.587953 | 0.103752 |
| Epi 1h | 790.716 | 240.027 | 0.645121 | 0.249618 |
| SA 1h | 1059.85 | 180.792 | 0.800725 | 0.135731 |
| Me-JA 1h | 872.971 | 184.864 | 0.818307 | 0.109114 |
| Control 6h | 2587.09 | 670.56 | 1.93414 | 0.356091 |
| ABA 6h | 1356.81 | 371.738 | 0.947289 | 0.305839 |
| ACC 6h | 1698.46 | 121.486 | 1.17414 | 0.15237 |
| BABA 6h | 1882.04 | 168.605 | 1.32924 | 0.130818 |
| Chitin 6h | 1959.29 | 326.16 | 1.4292 | 0.226189 |
| Epi 6h | 1381.12 | 79.9147 | 0.975915 | 0.096222 |
| SA 6h | 1481.76 | 376.252 | 1.12571 | 0.218163 |
| Me-JA 6h | 6117.35 | 1060.45 | 4.76377 | 0.529152 |
Source Transcript PGSC0003DMT400082012 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
