Probe CUST_27565_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27565_PI426222305 | JHI_St_60k_v1 | DMT400013620 | GGAACCCATTATGGGAAGATAGGCAACAGACTTCAAGAACTCAATGTATACCCTTAGGAG |
All Microarray Probes Designed to Gene DMG400005322
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27565_PI426222305 | JHI_St_60k_v1 | DMT400013620 | GGAACCCATTATGGGAAGATAGGCAACAGACTTCAAGAACTCAATGTATACCCTTAGGAG |
Microarray Signals from CUST_27565_PI426222305
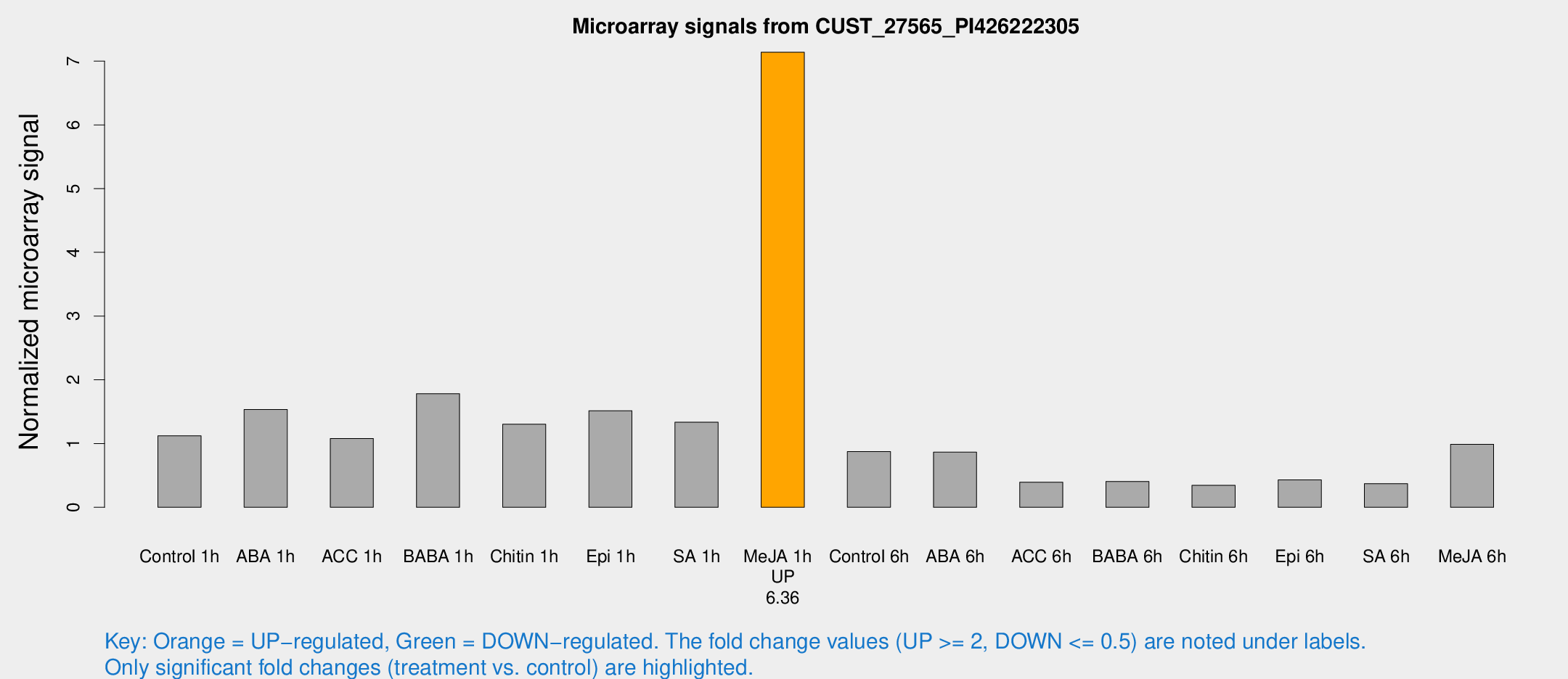
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 49.2067 | 5.49374 | 1.12269 | 0.104892 |
| ABA 1h | 60.3318 | 9.87551 | 1.53566 | 0.140977 |
| ACC 1h | 55.259 | 19.8183 | 1.07981 | 0.37467 |
| BABA 1h | 77.942 | 15.162 | 1.78339 | 0.209791 |
| Chitin 1h | 51.043 | 4.47485 | 1.30386 | 0.198938 |
| Epi 1h | 57.1665 | 4.73621 | 1.51596 | 0.127141 |
| SA 1h | 62.8716 | 15.2647 | 1.33472 | 0.304113 |
| Me-JA 1h | 261.851 | 51.1958 | 7.13954 | 0.751631 |
| Control 6h | 39.8466 | 8.43998 | 0.873192 | 0.134207 |
| ABA 6h | 42.3643 | 10.9916 | 0.865113 | 0.233765 |
| ACC 6h | 20.5168 | 4.5315 | 0.394735 | 0.102659 |
| BABA 6h | 19.7929 | 4.06603 | 0.405661 | 0.0840484 |
| Chitin 6h | 16.2007 | 4.12227 | 0.344427 | 0.0924354 |
| Epi 6h | 21.3863 | 4.36793 | 0.430689 | 0.0896295 |
| SA 6h | 16.7693 | 3.88456 | 0.371319 | 0.102328 |
| Me-JA 6h | 45.1203 | 10.2023 | 0.988512 | 0.195597 |
Source Transcript PGSC0003DMT400013620 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G60720.1 | +2 | 1e-19 | 85 | 45/137 (33%) | RNA-directed DNA polymerase (reverse transcriptase)-related family protein | chr1:22357072-22357941 FORWARD LENGTH=289 |
