Probe CUST_27495_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27495_PI426222305 | JHI_St_60k_v1 | DMT400024479 | AAATTAGAATGGACTGTGAAGGTTGTGCTCGTAAAGTCAAGAATGCTCTCTCTAAAGTAA |
All Microarray Probes Designed to Gene DMG400009466
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27495_PI426222305 | JHI_St_60k_v1 | DMT400024479 | AAATTAGAATGGACTGTGAAGGTTGTGCTCGTAAAGTCAAGAATGCTCTCTCTAAAGTAA |
| CUST_27460_PI426222305 | JHI_St_60k_v1 | DMT400024480 | TGTTGAAGAACAGGTAGGCTATGATACATGGACGTGCAAGTATTTTTATATGTTTCATAG |
Microarray Signals from CUST_27495_PI426222305
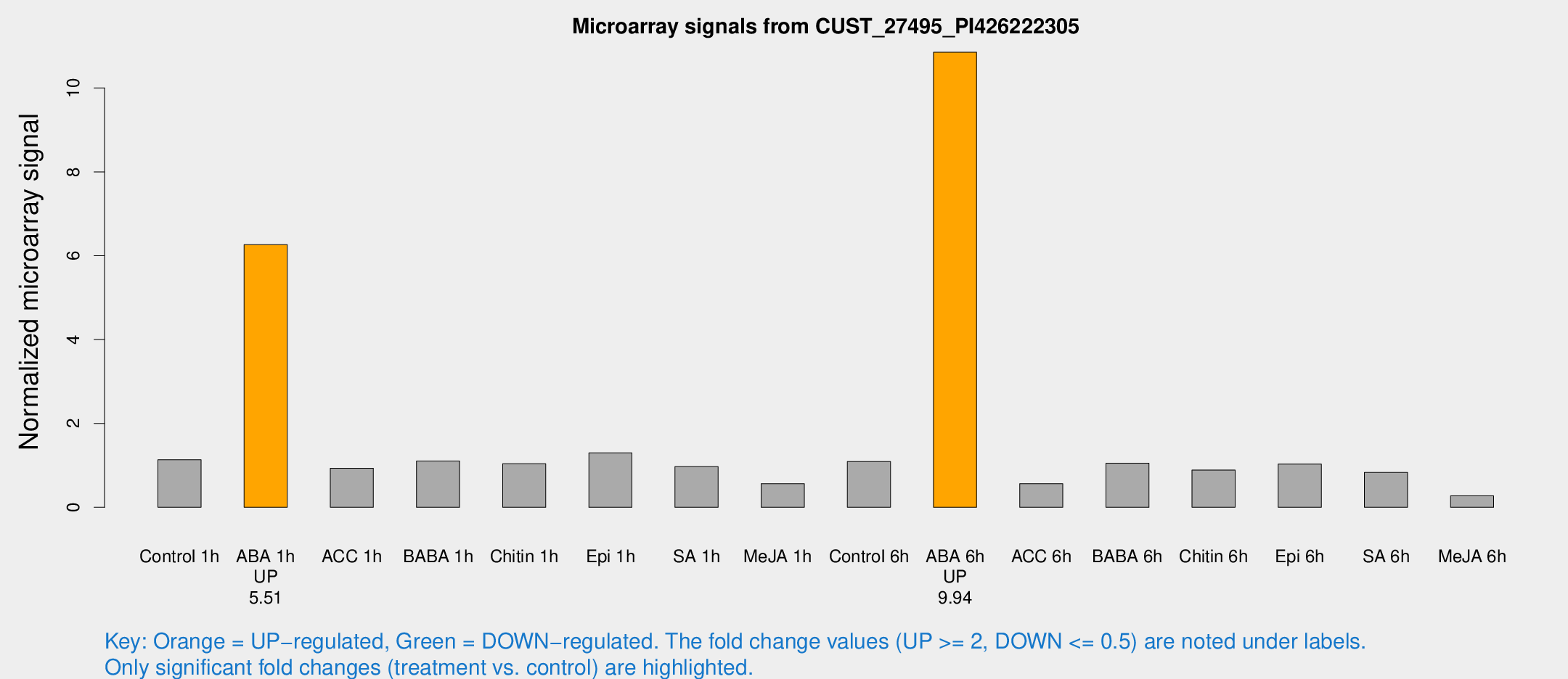
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 126.247 | 19.3527 | 1.13592 | 0.117607 |
| ABA 1h | 634.666 | 131.734 | 6.2639 | 1.14465 |
| ACC 1h | 110.535 | 25.124 | 0.931687 | 0.168907 |
| BABA 1h | 121.353 | 24.029 | 1.10438 | 0.135086 |
| Chitin 1h | 101.795 | 6.69033 | 1.03958 | 0.0681691 |
| Epi 1h | 123.245 | 11.5064 | 1.29784 | 0.0987867 |
| SA 1h | 114.736 | 29.1753 | 0.968873 | 0.182901 |
| Me-JA 1h | 50.6259 | 5.93823 | 0.562221 | 0.12076 |
| Control 6h | 120.709 | 14.2867 | 1.09173 | 0.0707696 |
| ABA 6h | 1284.63 | 191.424 | 10.8531 | 1.12881 |
| ACC 6h | 71.7821 | 10.4901 | 0.564021 | 0.045056 |
| BABA 6h | 129.515 | 13.2254 | 1.05281 | 0.0832478 |
| Chitin 6h | 103.149 | 7.02541 | 0.888654 | 0.074935 |
| Epi 6h | 130.479 | 22.6843 | 1.03272 | 0.227815 |
| SA 6h | 91.5742 | 13.5125 | 0.831728 | 0.0588014 |
| Me-JA 6h | 40.8498 | 21.9669 | 0.272382 | 0.16723 |
Source Transcript PGSC0003DMT400024479 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G08570.1 | +2 | 2e-54 | 177 | 89/151 (59%) | Heavy metal transport/detoxification superfamily protein | chr4:5455123-5455975 REVERSE LENGTH=150 |
