Probe CUST_27460_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27460_PI426222305 | JHI_St_60k_v1 | DMT400024480 | TGTTGAAGAACAGGTAGGCTATGATACATGGACGTGCAAGTATTTTTATATGTTTCATAG |
All Microarray Probes Designed to Gene DMG400009466
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27495_PI426222305 | JHI_St_60k_v1 | DMT400024479 | AAATTAGAATGGACTGTGAAGGTTGTGCTCGTAAAGTCAAGAATGCTCTCTCTAAAGTAA |
| CUST_27460_PI426222305 | JHI_St_60k_v1 | DMT400024480 | TGTTGAAGAACAGGTAGGCTATGATACATGGACGTGCAAGTATTTTTATATGTTTCATAG |
Microarray Signals from CUST_27460_PI426222305
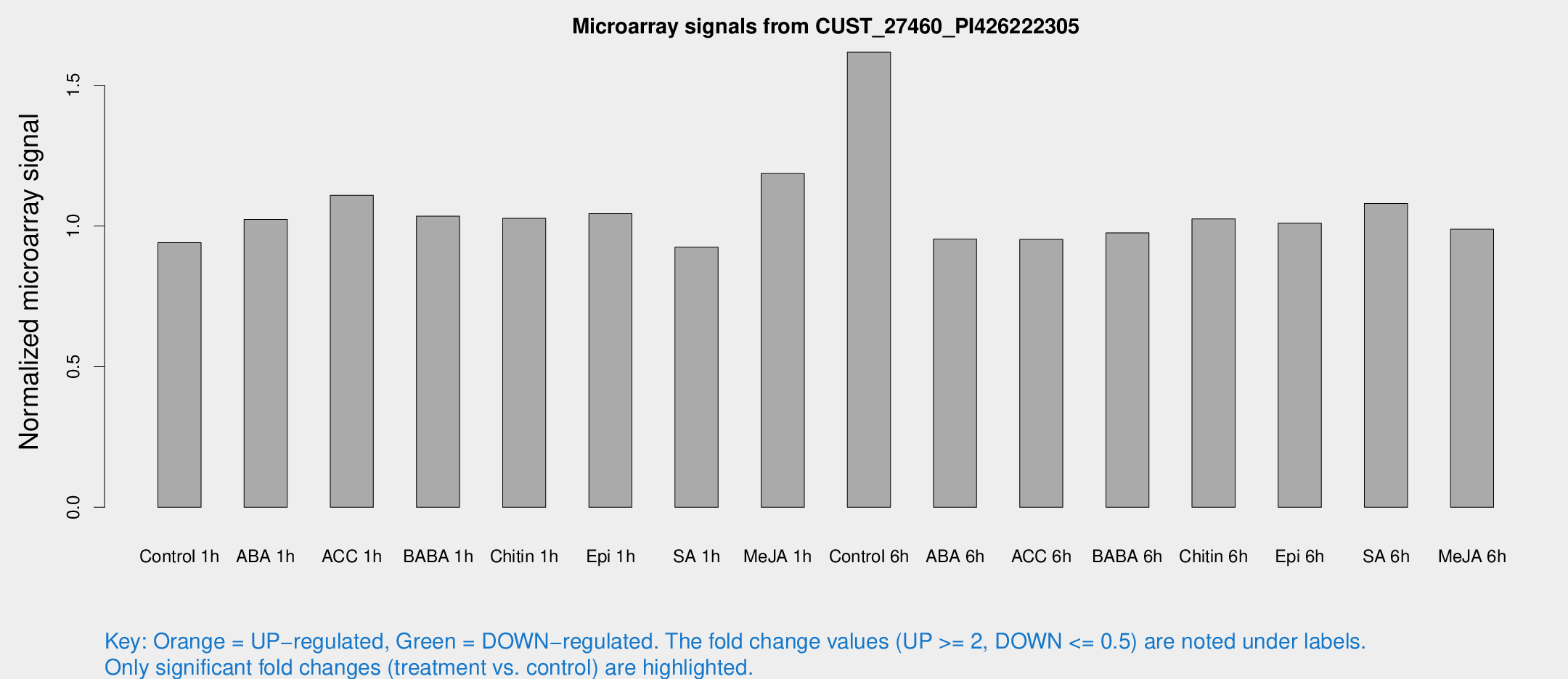
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.18763 | 3.59143 | 0.940759 | 0.544989 |
| ABA 1h | 5.94183 | 3.4423 | 1.02283 | 0.592329 |
| ACC 1h | 7.69166 | 4.59071 | 1.10851 | 0.641908 |
| BABA 1h | 6.56429 | 3.81408 | 1.0344 | 0.599263 |
| Chitin 1h | 6.10492 | 3.5599 | 1.02702 | 0.594733 |
| Epi 1h | 5.9543 | 3.46193 | 1.04363 | 0.604393 |
| SA 1h | 6.24024 | 3.61834 | 0.924183 | 0.53518 |
| Me-JA 1h | 6.39117 | 3.7225 | 1.18581 | 0.686597 |
| Control 6h | 11.8765 | 4.0384 | 1.61729 | 0.666817 |
| ABA 6h | 6.66061 | 3.862 | 0.953728 | 0.552649 |
| ACC 6h | 7.3344 | 4.35391 | 0.951925 | 0.551542 |
| BABA 6h | 7.18498 | 4.17208 | 0.975164 | 0.564699 |
| Chitin 6h | 7.16585 | 4.15831 | 1.02472 | 0.594301 |
| Epi 6h | 7.61304 | 4.49101 | 1.01045 | 0.585105 |
| SA 6h | 7.01479 | 4.0221 | 1.07982 | 0.619115 |
| Me-JA 6h | 6.46039 | 3.74107 | 0.98823 | 0.572244 |
Source Transcript PGSC0003DMT400024480 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G08570.1 | +3 | 5e-38 | 131 | 60/98 (61%) | Heavy metal transport/detoxification superfamily protein | chr4:5455123-5455975 REVERSE LENGTH=150 |
