Probe CUST_27422_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27422_PI426222305 | JHI_St_60k_v1 | DMT400070911 | TTGCTCTTCTGTTGCTCTTTTGGTTGACTACTGGATCTTCTAATTCAAGGTGTTTCTAAT |
All Microarray Probes Designed to Gene DMG400027574
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27422_PI426222305 | JHI_St_60k_v1 | DMT400070911 | TTGCTCTTCTGTTGCTCTTTTGGTTGACTACTGGATCTTCTAATTCAAGGTGTTTCTAAT |
Microarray Signals from CUST_27422_PI426222305
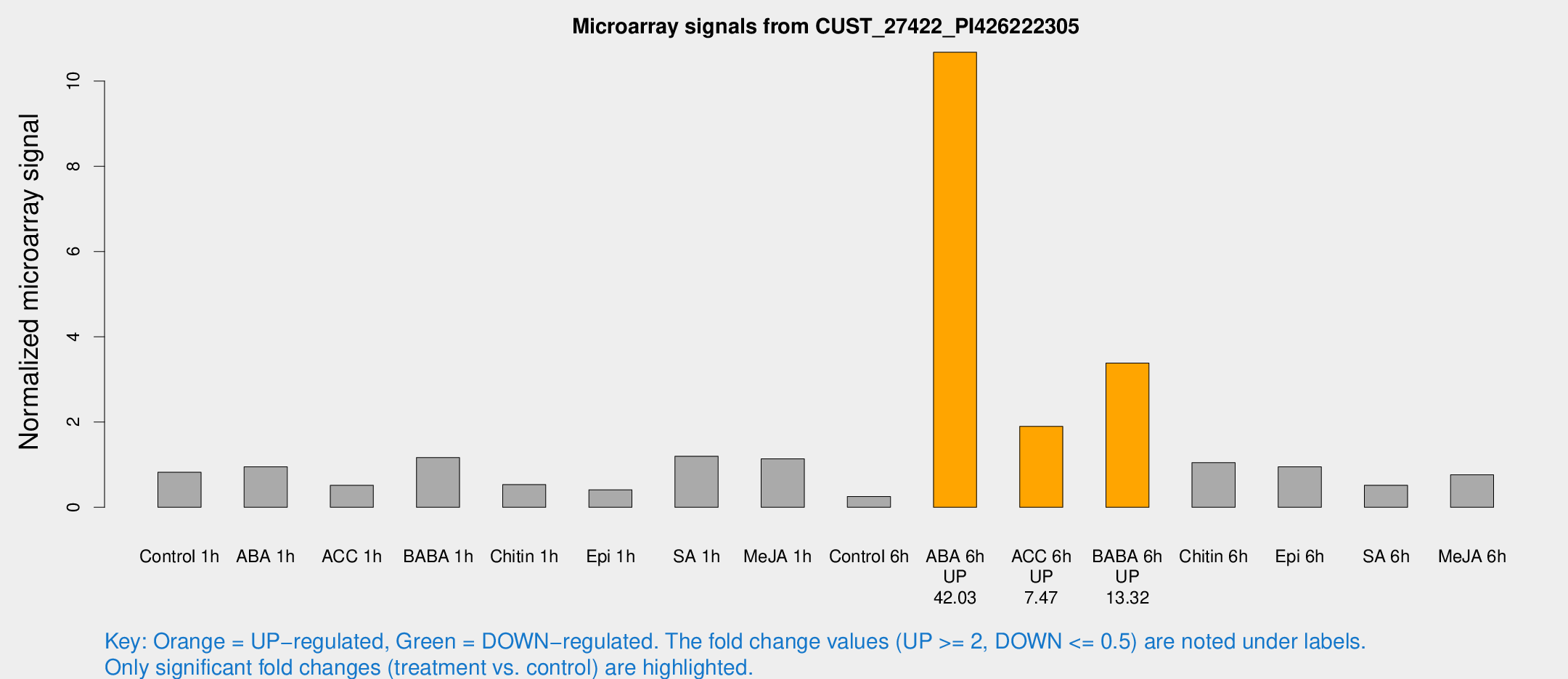
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 58.6081 | 17.4145 | 0.820947 | 0.23442 |
| ABA 1h | 69.5477 | 28.8866 | 0.948444 | 0.594532 |
| ACC 1h | 45.6915 | 18.4308 | 0.514477 | 0.356542 |
| BABA 1h | 78.6808 | 19.9474 | 1.16646 | 0.2428 |
| Chitin 1h | 41.364 | 20.7356 | 0.532756 | 0.353644 |
| Epi 1h | 23.6641 | 4.12361 | 0.411628 | 0.073795 |
| SA 1h | 81.4314 | 13.9568 | 1.19878 | 0.134285 |
| Me-JA 1h | 64.7479 | 19.3316 | 1.13712 | 0.235827 |
| Control 6h | 20.669 | 9.57634 | 0.253985 | 0.123584 |
| ABA 6h | 823.986 | 265.573 | 10.6743 | 3.51134 |
| ACC 6h | 140.178 | 9.25306 | 1.89804 | 0.273037 |
| BABA 6h | 262.44 | 74.9762 | 3.38237 | 0.818591 |
| Chitin 6h | 74.5237 | 15.0554 | 1.04741 | 0.233102 |
| Epi 6h | 69.8694 | 7.85058 | 0.951467 | 0.0826869 |
| SA 6h | 34.8949 | 8.64429 | 0.517639 | 0.0900885 |
| Me-JA 6h | 59.1072 | 21.4126 | 0.762012 | 0.435932 |
Source Transcript PGSC0003DMT400070911 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G63840.1 | +1 | 3e-16 | 73 | 29/54 (54%) | RING/U-box superfamily protein | chr1:23689991-23690491 REVERSE LENGTH=166 |
