Probe CUST_27235_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27235_PI426222305 | JHI_St_60k_v1 | DMT400040920 | TGATCACCTGTCATGTTTTCATTTCAGGAAGAAGCAGAATCGGAGTAAAGATTGGCATAG |
All Microarray Probes Designed to Gene DMG402015827
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27235_PI426222305 | JHI_St_60k_v1 | DMT400040920 | TGATCACCTGTCATGTTTTCATTTCAGGAAGAAGCAGAATCGGAGTAAAGATTGGCATAG |
Microarray Signals from CUST_27235_PI426222305
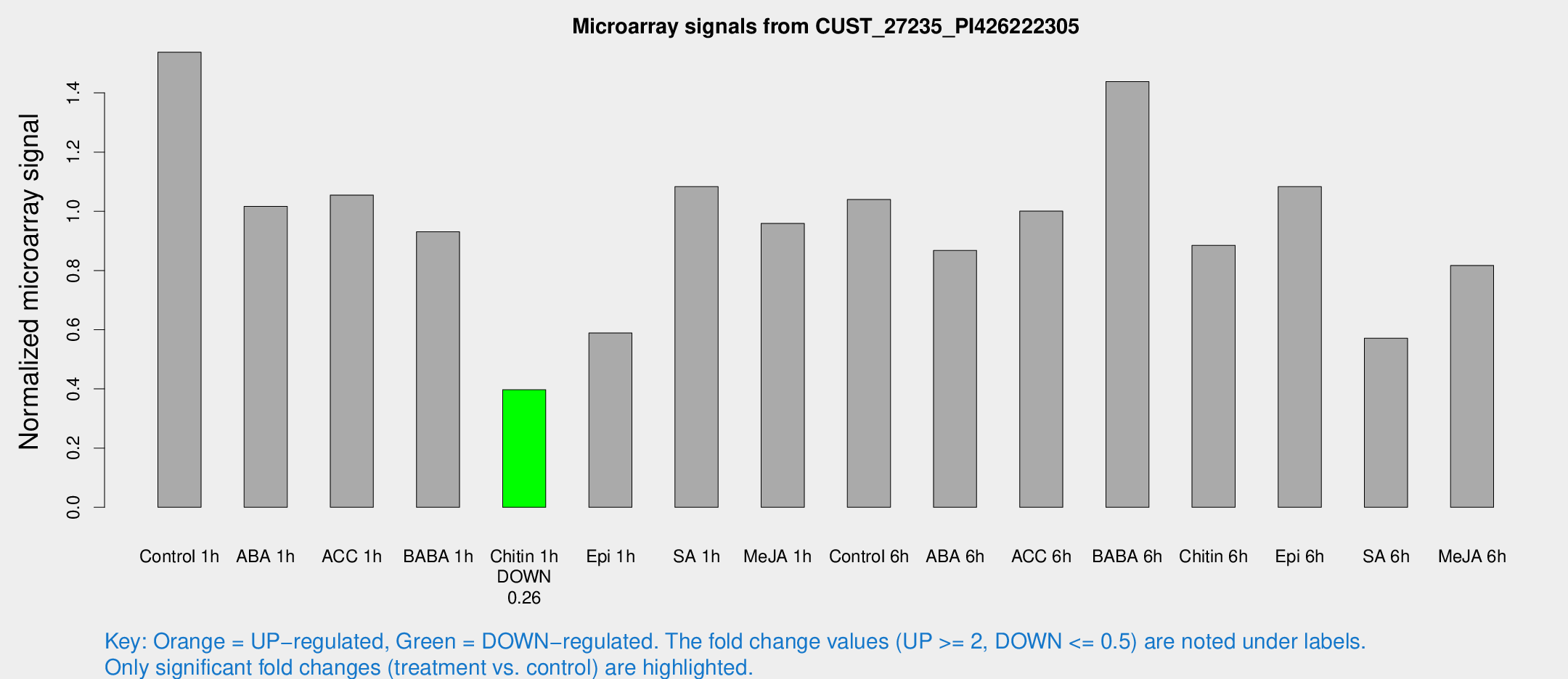
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 32.3495 | 4.05972 | 1.53744 | 0.194365 |
| ABA 1h | 19.2376 | 3.68757 | 1.01644 | 0.204058 |
| ACC 1h | 24.7396 | 7.34947 | 1.05487 | 0.335769 |
| BABA 1h | 23.4716 | 9.23896 | 0.930711 | 0.65264 |
| Chitin 1h | 7.63849 | 3.74223 | 0.397397 | 0.199565 |
| Epi 1h | 11.505 | 3.69326 | 0.588763 | 0.225646 |
| SA 1h | 23.3794 | 3.8401 | 1.08318 | 0.179636 |
| Me-JA 1h | 21.5983 | 10.1406 | 0.959051 | 0.62655 |
| Control 6h | 25.3728 | 8.87481 | 1.03981 | 0.350538 |
| ABA 6h | 21.0219 | 6.07311 | 0.867742 | 0.210238 |
| ACC 6h | 24.1734 | 4.82249 | 1.00046 | 0.210692 |
| BABA 6h | 35.2608 | 6.78914 | 1.43827 | 0.268319 |
| Chitin 6h | 20.7229 | 4.41378 | 0.885173 | 0.211936 |
| Epi 6h | 27.5686 | 7.55724 | 1.08345 | 0.348757 |
| SA 6h | 12.6013 | 4.13237 | 0.571325 | 0.214053 |
| Me-JA 6h | 22.0087 | 9.6451 | 0.816987 | 0.441309 |
Source Transcript PGSC0003DMT400040920 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G18390.2 | +3 | 3e-149 | 458 | 247/404 (61%) | Protein kinase superfamily protein | chr1:6327463-6329935 FORWARD LENGTH=654 |
