Probe CUST_27109_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27109_PI426222305 | JHI_St_60k_v1 | DMT400026276 | ATATCCTCGTCTGGCGACTGTCGATGACGATAAGGACTTTATTCCATTTGTGTTCATCAA |
All Microarray Probes Designed to Gene DMG400010139
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27109_PI426222305 | JHI_St_60k_v1 | DMT400026276 | ATATCCTCGTCTGGCGACTGTCGATGACGATAAGGACTTTATTCCATTTGTGTTCATCAA |
Microarray Signals from CUST_27109_PI426222305
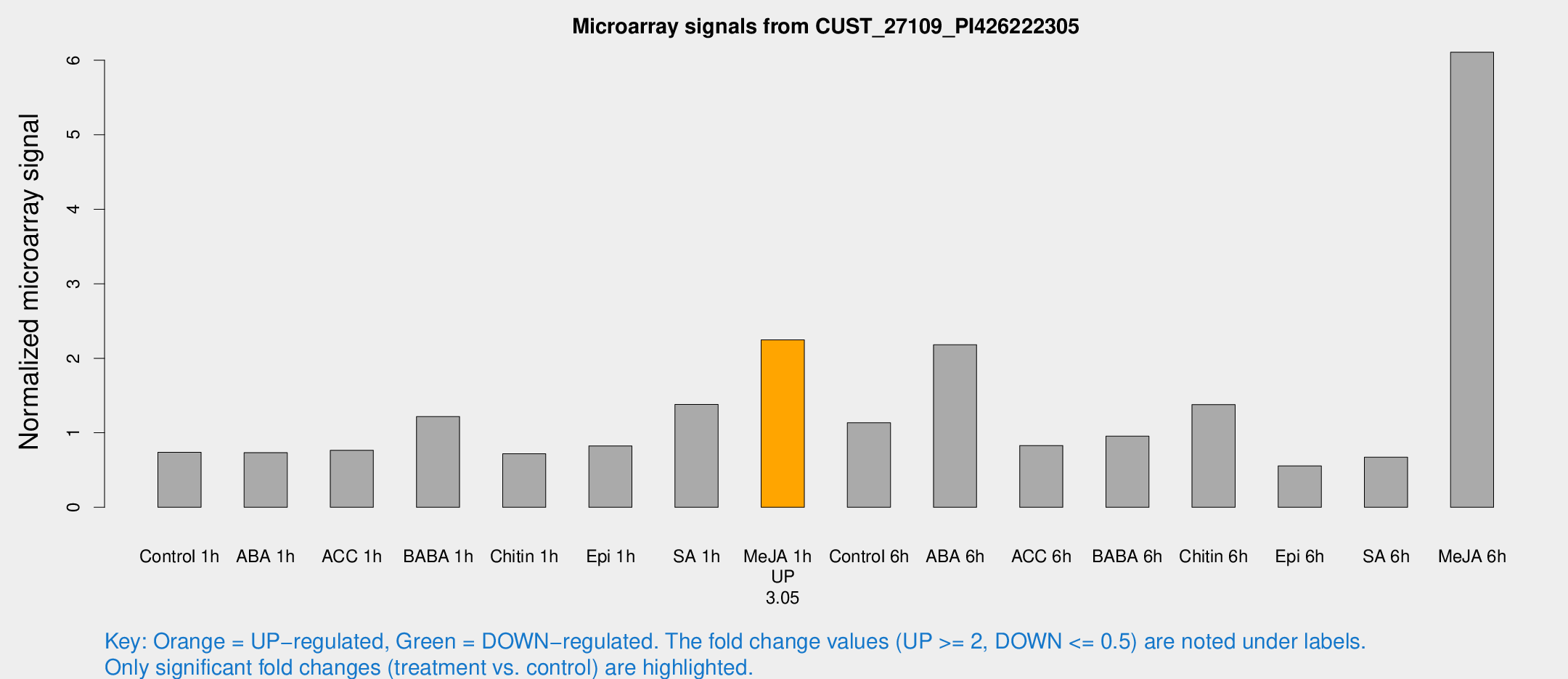
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7160.31 | 870.234 | 0.738132 | 0.111732 |
| ABA 1h | 7367.34 | 2796.2 | 0.732242 | 0.303431 |
| ACC 1h | 8770.15 | 2813.32 | 0.765199 | 0.299802 |
| BABA 1h | 11272.2 | 873.237 | 1.21763 | 0.0703008 |
| Chitin 1h | 6217.16 | 566.776 | 0.718562 | 0.0559918 |
| Epi 1h | 7248.57 | 1758.67 | 0.824161 | 0.215847 |
| SA 1h | 15218.9 | 4933.82 | 1.37967 | 0.608327 |
| Me-JA 1h | 17571.4 | 1017.72 | 2.2479 | 0.197604 |
| Control 6h | 14785.1 | 8335.95 | 1.13401 | 0.638358 |
| ABA 6h | 22291.9 | 1871.17 | 2.18113 | 0.254759 |
| ACC 6h | 10295.8 | 3089.5 | 0.8288 | 0.310165 |
| BABA 6h | 10246.1 | 748.082 | 0.953803 | 0.055069 |
| Chitin 6h | 15932.6 | 5693.99 | 1.37909 | 0.574657 |
| Epi 6h | 8573.04 | 4227.41 | 0.556149 | 0.652508 |
| SA 6h | 6384.65 | 478.436 | 0.672484 | 0.102608 |
| Me-JA 6h | 60732.1 | 11819.8 | 6.10682 | 0.887646 |
Source Transcript PGSC0003DMT400026276 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73325.1 | +1 | 5e-06 | 46 | 49/181 (27%) | Kunitz family trypsin and protease inhibitor protein | chr1:27567518-27568186 REVERSE LENGTH=222 |
