Probe CUST_27047_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27047_PI426222305 | JHI_St_60k_v1 | DMT400026280 | GGCTCTCAATAACAAGCCTTATCCATTTGGCTTCACCAAAGTAAACAAGAATATTGATGC |
All Microarray Probes Designed to Gene DMG400010141
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27047_PI426222305 | JHI_St_60k_v1 | DMT400026280 | GGCTCTCAATAACAAGCCTTATCCATTTGGCTTCACCAAAGTAAACAAGAATATTGATGC |
Microarray Signals from CUST_27047_PI426222305
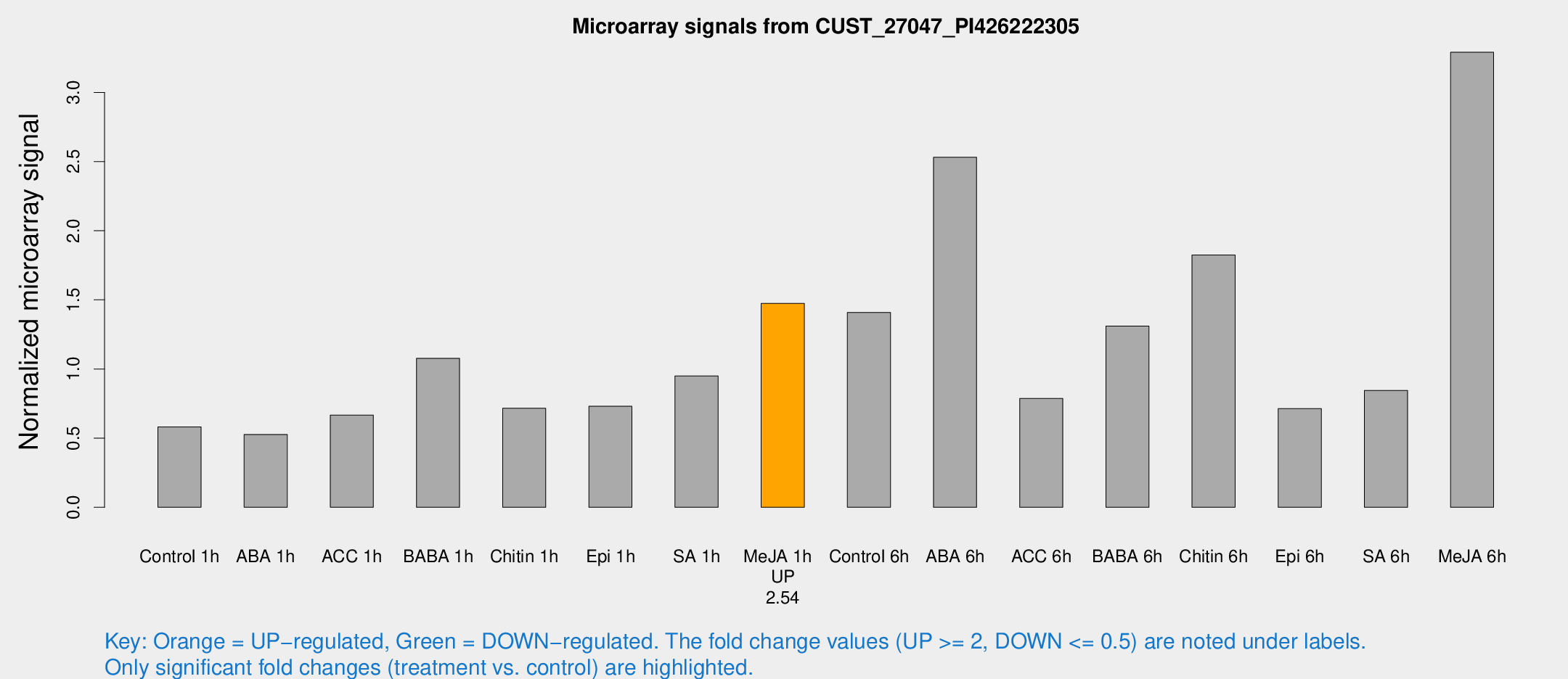
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2192.69 | 126.981 | 0.581789 | 0.0335987 |
| ABA 1h | 2245.22 | 926.567 | 0.526657 | 0.304739 |
| ACC 1h | 2911.48 | 864.893 | 0.666382 | 0.214443 |
| BABA 1h | 3973.89 | 530.276 | 1.07737 | 0.0752323 |
| Chitin 1h | 2474.18 | 383.665 | 0.715928 | 0.0609032 |
| Epi 1h | 2577.42 | 648.156 | 0.73046 | 0.220261 |
| SA 1h | 3829.06 | 835.982 | 0.94934 | 0.21435 |
| Me-JA 1h | 4714.11 | 982.133 | 1.47484 | 0.22504 |
| Control 6h | 6998.86 | 3796.95 | 1.40896 | 0.742199 |
| ABA 6h | 10702 | 2567.29 | 2.53119 | 0.725314 |
| ACC 6h | 3645.3 | 929.07 | 0.786819 | 0.165709 |
| BABA 6h | 5568.42 | 579.365 | 1.31001 | 0.101794 |
| Chitin 6h | 8366.02 | 3245.49 | 1.82435 | 0.730141 |
| Epi 6h | 3348.54 | 1009.42 | 0.713237 | 0.328544 |
| SA 6h | 3170.37 | 305.244 | 0.845108 | 0.120141 |
| Me-JA 6h | 13013.7 | 3178.36 | 3.29069 | 0.524013 |
Source Transcript PGSC0003DMT400026280 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73260.1 | +1 | 2e-21 | 89 | 64/188 (34%) | kunitz trypsin inhibitor 1 | chr1:27547410-27548057 REVERSE LENGTH=215 |
