Probe CUST_27028_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27028_PI426222305 | JHI_St_60k_v1 | DMT400052687 | GTGCAGCTAAAGAGAAAGATCTCTACTTAATTGCCAAAACAAATCAAAGTTTGGATGTCA |
All Microarray Probes Designed to Gene DMG403020453
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27028_PI426222305 | JHI_St_60k_v1 | DMT400052687 | GTGCAGCTAAAGAGAAAGATCTCTACTTAATTGCCAAAACAAATCAAAGTTTGGATGTCA |
Microarray Signals from CUST_27028_PI426222305
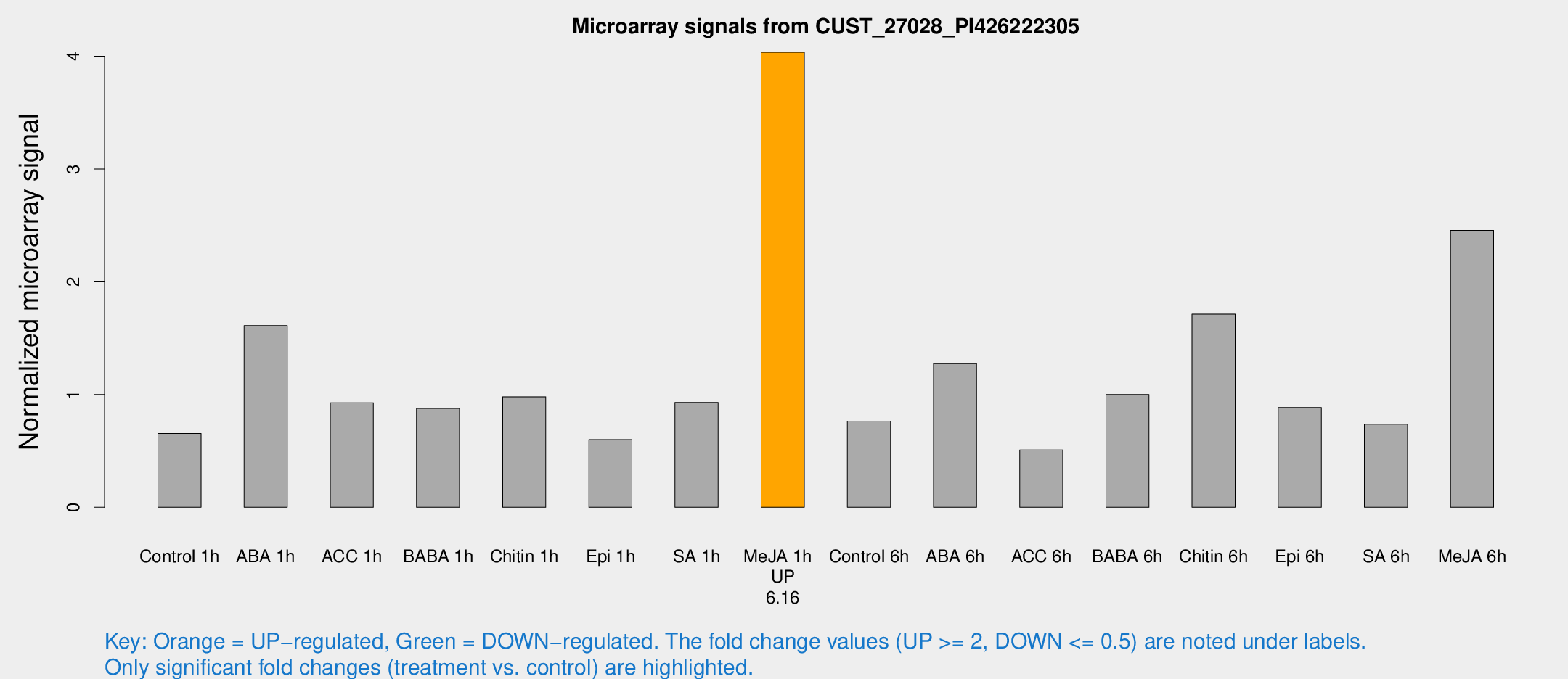
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14.0696 | 5.41896 | 0.654968 | 0.311833 |
| ABA 1h | 26.3087 | 4.30412 | 1.61136 | 0.206481 |
| ACC 1h | 17.8296 | 3.69342 | 0.927232 | 0.204001 |
| BABA 1h | 16.39 | 4.5059 | 0.877657 | 0.205695 |
| Chitin 1h | 15.8876 | 3.10426 | 0.980178 | 0.190477 |
| Epi 1h | 10.0282 | 2.9953 | 0.599799 | 0.218993 |
| SA 1h | 17.2554 | 3.13984 | 0.929447 | 0.170564 |
| Me-JA 1h | 62.4553 | 14.128 | 4.03535 | 0.575596 |
| Control 6h | 19.0249 | 10.3157 | 0.764418 | 0.599839 |
| ABA 6h | 25.7326 | 5.68095 | 1.27421 | 0.290159 |
| ACC 6h | 10.9538 | 3.71245 | 0.508651 | 0.1953 |
| BABA 6h | 20.6448 | 3.59936 | 1.00049 | 0.184441 |
| Chitin 6h | 33.0176 | 3.86831 | 1.71325 | 0.202992 |
| Epi 6h | 18.0518 | 3.67084 | 0.884814 | 0.18528 |
| SA 6h | 15.6861 | 5.85936 | 0.736731 | 0.429554 |
| Me-JA 6h | 46.5589 | 10.0539 | 2.45634 | 0.408638 |
Source Transcript PGSC0003DMT400052687 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G26210.1 | +1 | 2e-58 | 194 | 100/204 (49%) | cytochrome P450, family 71, subfamily B, polypeptide 23 | chr3:9593329-9595202 REVERSE LENGTH=501 |
