Probe CUST_26713_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26713_PI426222305 | JHI_St_60k_v1 | DMT400061524 | AACCCTAAATCCTAATGCTCCAATGTTCGTTCCCTCCGAATATCGTCAAGTGGAGGACTT |
All Microarray Probes Designed to Gene DMG400023945
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26713_PI426222305 | JHI_St_60k_v1 | DMT400061524 | AACCCTAAATCCTAATGCTCCAATGTTCGTTCCCTCCGAATATCGTCAAGTGGAGGACTT |
Microarray Signals from CUST_26713_PI426222305
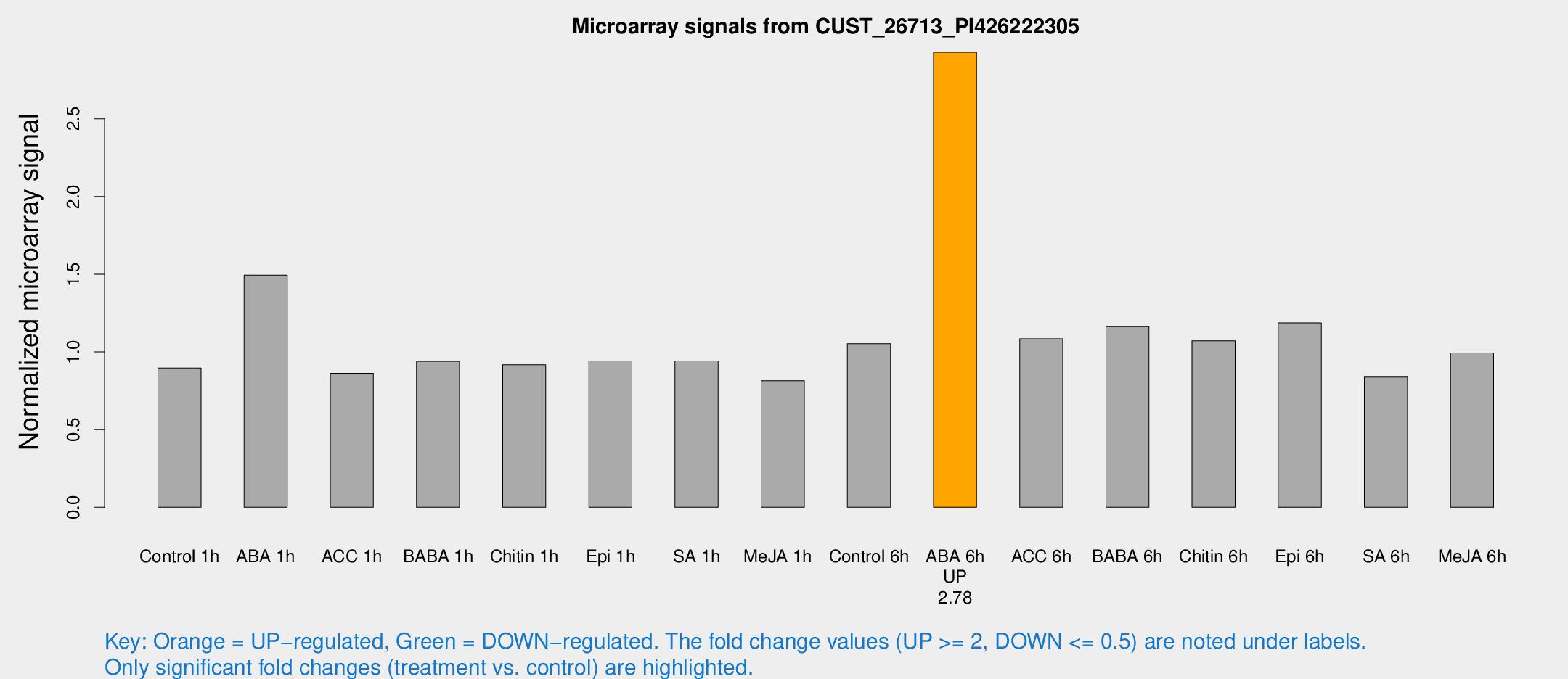
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 215.169 | 15.8407 | 0.896316 | 0.0531182 |
| ABA 1h | 329.249 | 67.7953 | 1.49459 | 0.20362 |
| ACC 1h | 214.409 | 23.2933 | 0.863351 | 0.051599 |
| BABA 1h | 218.804 | 24.3727 | 0.939411 | 0.0791874 |
| Chitin 1h | 197.797 | 12.7279 | 0.917335 | 0.0659534 |
| Epi 1h | 195.843 | 14.9779 | 0.941701 | 0.0623452 |
| SA 1h | 234.016 | 27.3079 | 0.941842 | 0.0618002 |
| Me-JA 1h | 159.326 | 9.66929 | 0.815248 | 0.0493662 |
| Control 6h | 264.291 | 54.4573 | 1.0533 | 0.154704 |
| ABA 6h | 748.788 | 61.7226 | 2.92834 | 0.175056 |
| ACC 6h | 307.68 | 59.8142 | 1.08489 | 0.0638736 |
| BABA 6h | 316.456 | 42.7676 | 1.16235 | 0.112833 |
| Chitin 6h | 274.613 | 25.1146 | 1.07177 | 0.125657 |
| Epi 6h | 321.082 | 21.6788 | 1.18704 | 0.147351 |
| SA 6h | 210.061 | 49.734 | 0.838292 | 0.251572 |
| Me-JA 6h | 243.623 | 43.3436 | 0.993871 | 0.0943166 |
Source Transcript PGSC0003DMT400061524 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G14270.1 | +1 | 2e-15 | 74 | 52/137 (38%) | Protein containing PAM2 motif which mediates interaction with the PABC domain of polyadenyl binding proteins. | chr4:8218517-8219037 FORWARD LENGTH=143 |
