Probe CUST_26671_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26671_PI426222305 | JHI_St_60k_v1 | DMT400000736 | TATTAATGCTGTTCAATCTGTTATTCAAGCTTATCCTTCTTGAATGGCTGCTCTCCTGCT |
All Microarray Probes Designed to Gene DMG400000270
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26671_PI426222305 | JHI_St_60k_v1 | DMT400000736 | TATTAATGCTGTTCAATCTGTTATTCAAGCTTATCCTTCTTGAATGGCTGCTCTCCTGCT |
Microarray Signals from CUST_26671_PI426222305
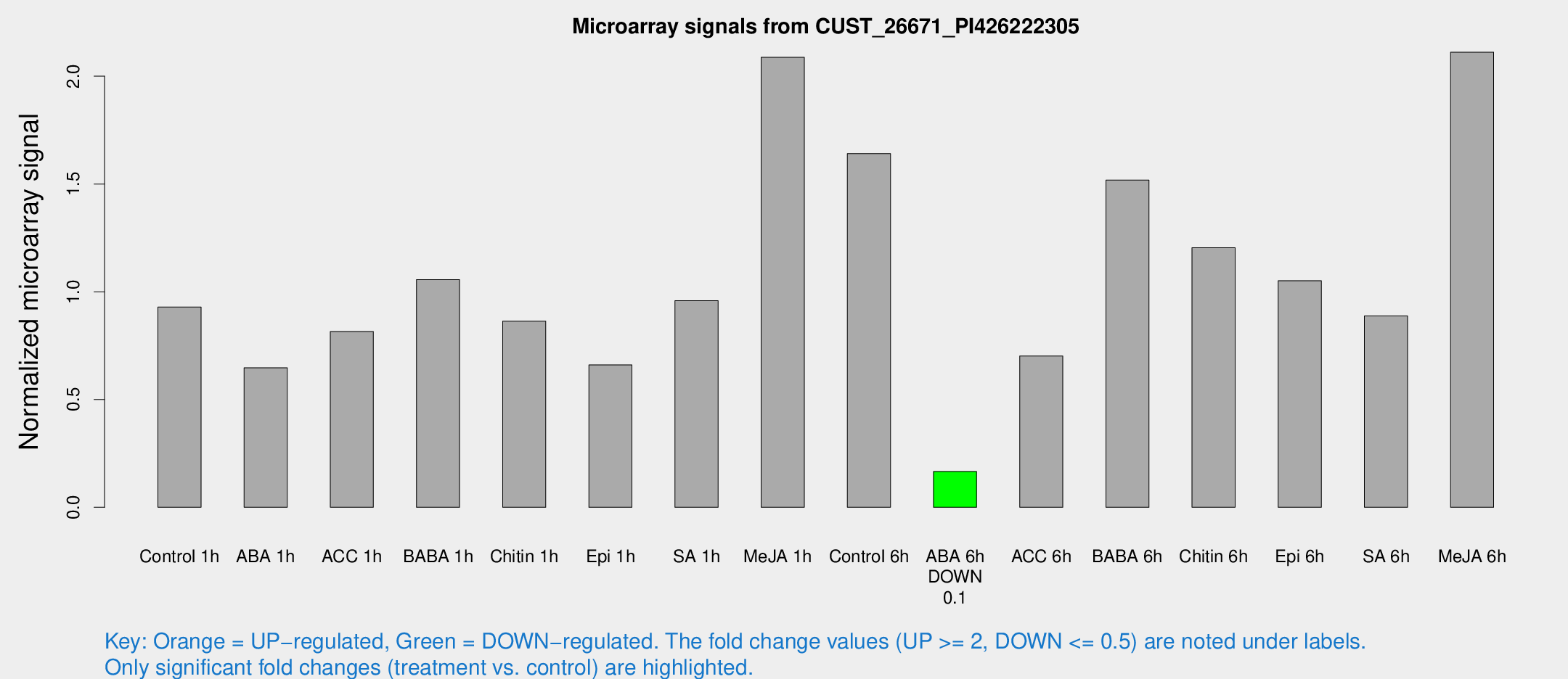
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 418.512 | 63.7253 | 0.928957 | 0.120275 |
| ABA 1h | 256.815 | 35.9023 | 0.647052 | 0.0690817 |
| ACC 1h | 380.322 | 68.1374 | 0.815324 | 0.107791 |
| BABA 1h | 456.052 | 58.9591 | 1.05649 | 0.061545 |
| Chitin 1h | 353.048 | 64.9444 | 0.863757 | 0.152228 |
| Epi 1h | 253.184 | 19.7248 | 0.66063 | 0.0390632 |
| SA 1h | 509.432 | 201.419 | 0.958818 | 0.480807 |
| Me-JA 1h | 771.097 | 127.819 | 2.08751 | 0.314272 |
| Control 6h | 840.961 | 268.605 | 1.64122 | 0.579508 |
| ABA 6h | 79.4169 | 10.9843 | 0.166285 | 0.0223344 |
| ACC 6h | 362.732 | 56.9188 | 0.70191 | 0.0412182 |
| BABA 6h | 760.041 | 91.2185 | 1.51802 | 0.222713 |
| Chitin 6h | 572.433 | 65.7919 | 1.20396 | 0.118654 |
| Epi 6h | 527.298 | 52.773 | 1.05139 | 0.16872 |
| SA 6h | 406.976 | 87.3893 | 0.888056 | 0.115135 |
| Me-JA 6h | 954.562 | 166.884 | 2.1114 | 0.489049 |
Source Transcript PGSC0003DMT400000736 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G35160.1 | +1 | 1e-79 | 248 | 136/255 (53%) | O-methyltransferase family protein | chr4:16730989-16732808 REVERSE LENGTH=382 |
