Probe CUST_26567_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26567_PI426222305 | JHI_St_60k_v1 | DMT400000810 | TAAGGATGGATGAGTACGTACCATCTGATTCGTGATAGTAAAAGGCTTTTACTTACTTTC |
All Microarray Probes Designed to Gene DMG400000301
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26567_PI426222305 | JHI_St_60k_v1 | DMT400000810 | TAAGGATGGATGAGTACGTACCATCTGATTCGTGATAGTAAAAGGCTTTTACTTACTTTC |
Microarray Signals from CUST_26567_PI426222305
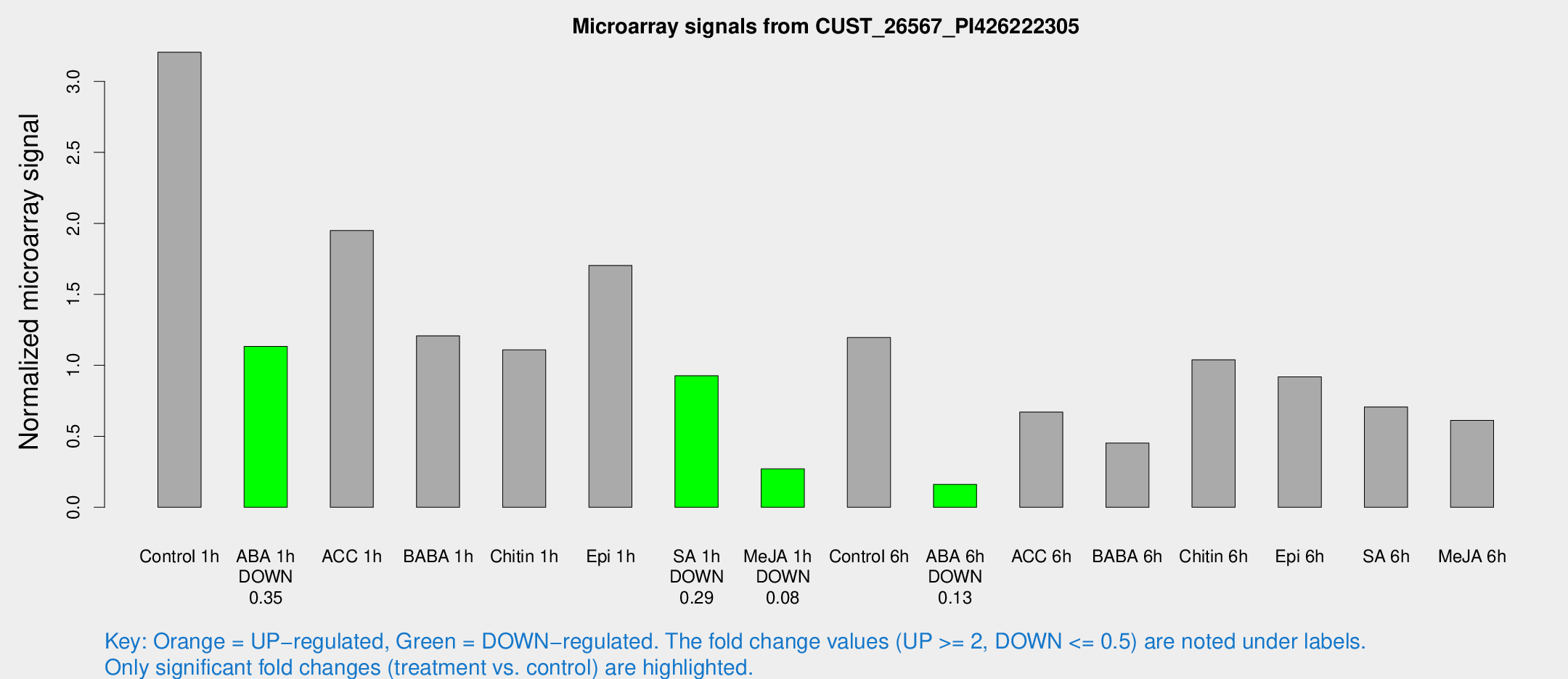
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 103.093 | 10.2716 | 3.2054 | 0.206981 |
| ABA 1h | 33.0027 | 6.16937 | 1.13395 | 0.278608 |
| ACC 1h | 65.9344 | 11.1819 | 1.94972 | 0.447005 |
| BABA 1h | 42.8671 | 13.4679 | 1.20778 | 0.401362 |
| Chitin 1h | 36.1009 | 12.4833 | 1.10921 | 0.334151 |
| Epi 1h | 46.9846 | 3.99363 | 1.7037 | 0.144819 |
| SA 1h | 31.5927 | 6.1273 | 0.926669 | 0.185044 |
| Me-JA 1h | 7.42018 | 3.02144 | 0.271222 | 0.120242 |
| Control 6h | 46.699 | 19.4572 | 1.19629 | 0.525891 |
| ABA 6h | 5.45475 | 3.16707 | 0.160843 | 0.0932779 |
| ACC 6h | 25.4712 | 4.55679 | 0.671012 | 0.113357 |
| BABA 6h | 19.9039 | 9.23994 | 0.452564 | 0.226267 |
| Chitin 6h | 37.8305 | 10.4134 | 1.03868 | 0.249666 |
| Epi 6h | 36.6209 | 10.1684 | 0.919319 | 0.433393 |
| SA 6h | 31.0503 | 12.655 | 0.706261 | 0.500858 |
| Me-JA 6h | 21.5504 | 6.66218 | 0.612796 | 0.15475 |
Source Transcript PGSC0003DMT400000810 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G28890.1 | +2 | 9e-92 | 289 | 172/397 (43%) | RING/U-box superfamily protein | chr4:14256437-14257735 REVERSE LENGTH=432 |
