Probe CUST_26407_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26407_PI426222305 | JHI_St_60k_v1 | DMT400037207 | TTCATATCACTCATGCACGGCTTCACGAACTTGTTGAACGTGCCGCTTCTCAACTTGTAT |
All Microarray Probes Designed to Gene DMG400014345
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26407_PI426222305 | JHI_St_60k_v1 | DMT400037207 | TTCATATCACTCATGCACGGCTTCACGAACTTGTTGAACGTGCCGCTTCTCAACTTGTAT |
Microarray Signals from CUST_26407_PI426222305
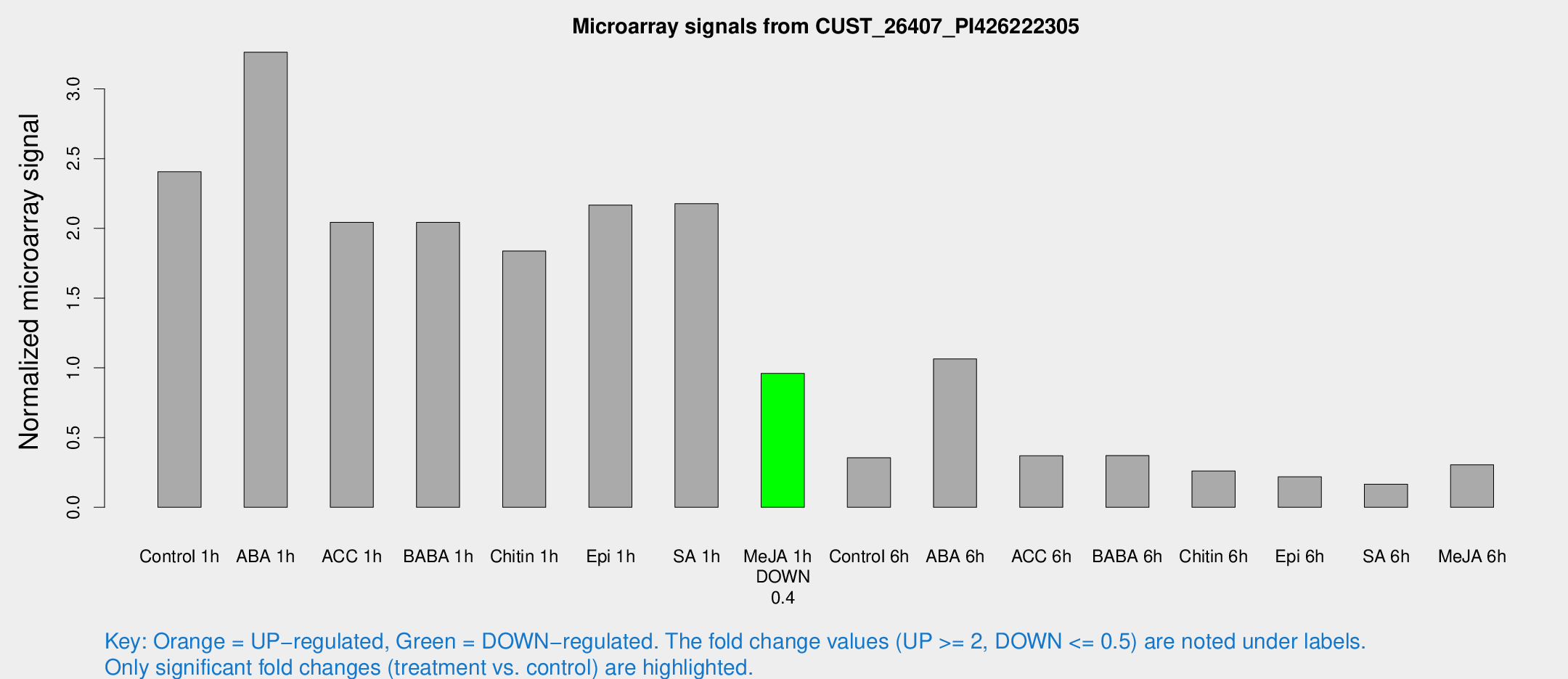
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 332.512 | 47.6995 | 2.40559 | 0.283942 |
| ABA 1h | 399.531 | 60.7644 | 3.26258 | 0.44166 |
| ACC 1h | 330.901 | 108.784 | 2.04299 | 0.778008 |
| BABA 1h | 279.48 | 56.3961 | 2.04322 | 0.258438 |
| Chitin 1h | 226.075 | 24.3341 | 1.83697 | 0.109435 |
| Epi 1h | 253.529 | 14.9844 | 2.16712 | 0.128078 |
| SA 1h | 331.437 | 88.6777 | 2.17642 | 0.606791 |
| Me-JA 1h | 106.412 | 7.54536 | 0.959426 | 0.0631362 |
| Control 6h | 49.845 | 8.88472 | 0.35503 | 0.040121 |
| ABA 6h | 177.233 | 59.1774 | 1.06335 | 0.448326 |
| ACC 6h | 58.0414 | 6.48484 | 0.369546 | 0.068348 |
| BABA 6h | 56.1903 | 5.0506 | 0.370361 | 0.0331679 |
| Chitin 6h | 37.6185 | 4.41971 | 0.260644 | 0.0309174 |
| Epi 6h | 34.0585 | 5.30729 | 0.21833 | 0.0414218 |
| SA 6h | 23.2159 | 4.8151 | 0.165073 | 0.0589384 |
| Me-JA 6h | 43.6636 | 11.4 | 0.304875 | 0.053917 |
Source Transcript PGSC0003DMT400037207 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G48990.1 | +3 | 0.0 | 795 | 403/511 (79%) | AMP-dependent synthetase and ligase family protein | chr3:18159031-18161294 REVERSE LENGTH=514 |
