Probe CUST_26158_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26158_PI426222305 | JHI_St_60k_v1 | DMT400041634 | GTGTTCTACTATGAGATTCTGAATGCATCAGAAAAAGCATGCAGCATGGCTAAGCAGGCA |
All Microarray Probes Designed to Gene DMG401016141
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26133_PI426222305 | JHI_St_60k_v1 | DMT400041636 | TGGAGTGCCCTTAGAAGAAGGAACCTTTCCTCTCCTAGTTGTTTCATAGTCGTTTTGCTT |
| CUST_26158_PI426222305 | JHI_St_60k_v1 | DMT400041634 | GTGTTCTACTATGAGATTCTGAATGCATCAGAAAAAGCATGCAGCATGGCTAAGCAGGCA |
Microarray Signals from CUST_26158_PI426222305
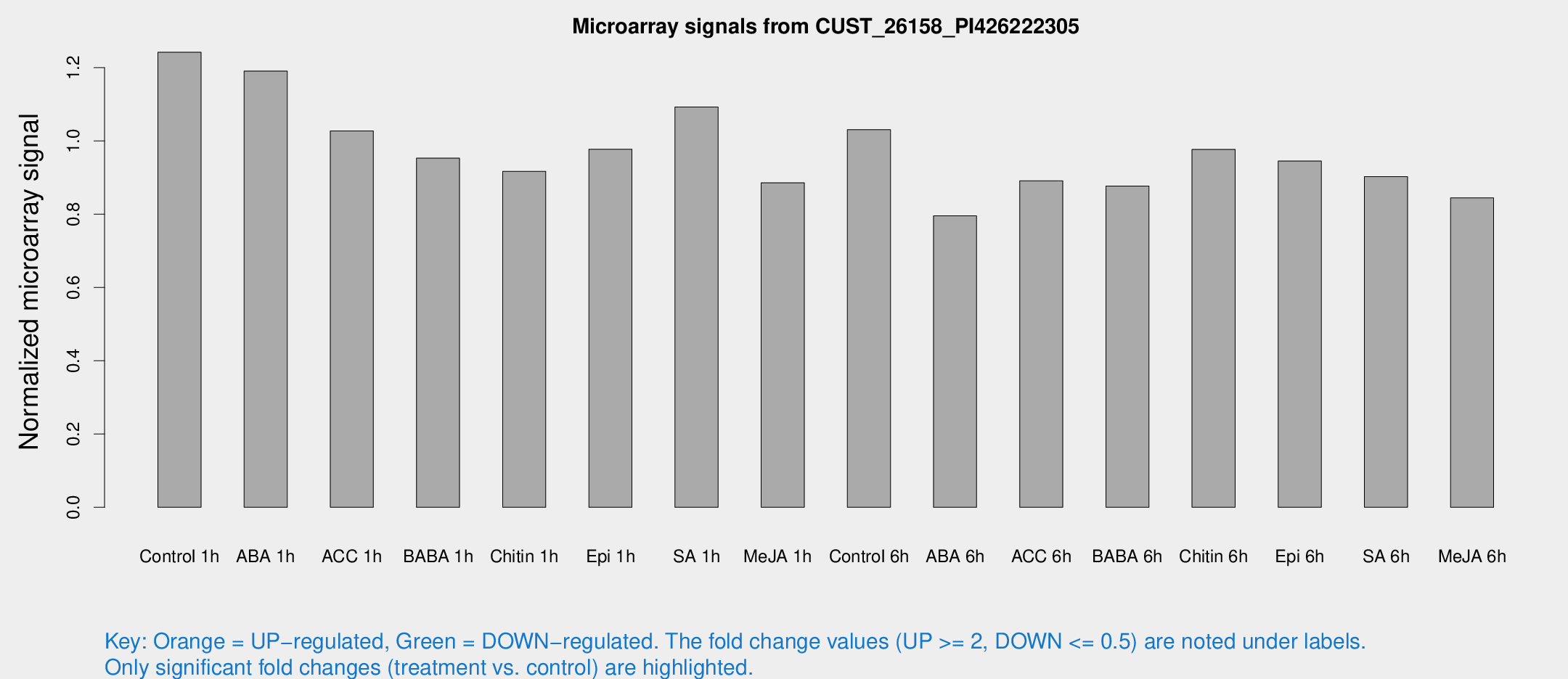
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 12379.9 | 1051.58 | 1.24199 | 0.0717072 |
| ABA 1h | 10540.9 | 1162.97 | 1.19059 | 0.0687394 |
| ACC 1h | 10801.8 | 1849.99 | 1.02732 | 0.117775 |
| BABA 1h | 9735.31 | 2319.76 | 0.952905 | 0.167207 |
| Chitin 1h | 8320.31 | 1185.82 | 0.916915 | 0.0568991 |
| Epi 1h | 8384.97 | 485.085 | 0.977349 | 0.0564285 |
| SA 1h | 11499.9 | 2001.91 | 1.09232 | 0.154385 |
| Me-JA 1h | 7257.04 | 844.646 | 0.885384 | 0.0511192 |
| Control 6h | 10695.2 | 2115.53 | 1.03076 | 0.141092 |
| ABA 6h | 8475.22 | 952.944 | 0.795348 | 0.0491125 |
| ACC 6h | 10226.5 | 1068.57 | 0.891001 | 0.061361 |
| BABA 6h | 9890.2 | 1364.91 | 0.876893 | 0.105212 |
| Chitin 6h | 10342.4 | 759.57 | 0.976838 | 0.0614181 |
| Epi 6h | 10588.9 | 710.743 | 0.945226 | 0.090949 |
| SA 6h | 9320.22 | 1949.69 | 0.902475 | 0.112897 |
| Me-JA 6h | 8542.25 | 1423.66 | 0.844418 | 0.0717721 |
Source Transcript PGSC0003DMT400041634 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65430.3 | +2 | 7e-139 | 400 | 203/238 (85%) | general regulatory factor 8 | chr5:26148630-26150255 REVERSE LENGTH=260 |
