Probe CUST_26133_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26133_PI426222305 | JHI_St_60k_v1 | DMT400041636 | TGGAGTGCCCTTAGAAGAAGGAACCTTTCCTCTCCTAGTTGTTTCATAGTCGTTTTGCTT |
All Microarray Probes Designed to Gene DMG401016141
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26158_PI426222305 | JHI_St_60k_v1 | DMT400041634 | GTGTTCTACTATGAGATTCTGAATGCATCAGAAAAAGCATGCAGCATGGCTAAGCAGGCA |
| CUST_26133_PI426222305 | JHI_St_60k_v1 | DMT400041636 | TGGAGTGCCCTTAGAAGAAGGAACCTTTCCTCTCCTAGTTGTTTCATAGTCGTTTTGCTT |
Microarray Signals from CUST_26133_PI426222305
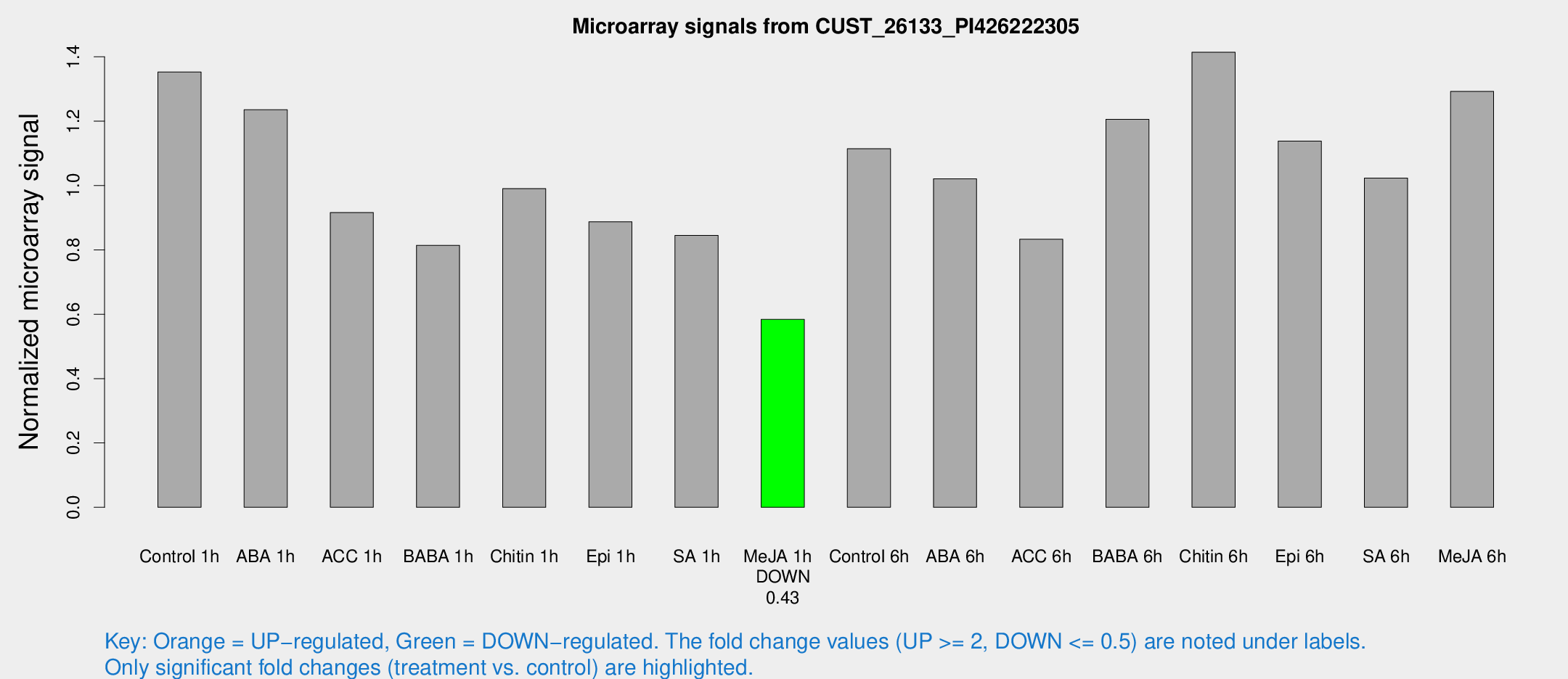
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 69.9456 | 5.22623 | 1.35304 | 0.0954614 |
| ABA 1h | 56.4139 | 4.24592 | 1.23567 | 0.131438 |
| ACC 1h | 49.9148 | 8.66999 | 0.915995 | 0.0988141 |
| BABA 1h | 42.8952 | 9.91158 | 0.814293 | 0.130228 |
| Chitin 1h | 45.9236 | 3.87684 | 0.990411 | 0.137591 |
| Epi 1h | 39.6096 | 3.62699 | 0.887546 | 0.0818561 |
| SA 1h | 44.973 | 3.99464 | 0.844938 | 0.0732948 |
| Me-JA 1h | 24.8137 | 3.22661 | 0.584065 | 0.0774764 |
| Control 6h | 62.4242 | 18.1123 | 1.11427 | 0.26576 |
| ABA 6h | 56.7626 | 7.07835 | 1.02111 | 0.0911157 |
| ACC 6h | 51.7479 | 10.8896 | 0.833047 | 0.170144 |
| BABA 6h | 73.4395 | 18.394 | 1.20592 | 0.286416 |
| Chitin 6h | 77.5665 | 5.53327 | 1.41433 | 0.104094 |
| Epi 6h | 68.4479 | 12.7113 | 1.13834 | 0.290558 |
| SA 6h | 60.7302 | 24.2118 | 1.02324 | 0.438255 |
| Me-JA 6h | 69.0667 | 13.3757 | 1.29256 | 0.203424 |
Source Transcript PGSC0003DMT400041636 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G65430.3 | +3 | 1e-18 | 85 | 44/50 (88%) | general regulatory factor 8 | chr5:26148630-26150255 REVERSE LENGTH=260 |
