Probe CUST_26092_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26092_PI426222305 | JHI_St_60k_v1 | DMT400041820 | CGAAGATGTAAATGTGATTTCTGTGTTCATCAGATTATATGTACTCCGCGGAATCATATA |
All Microarray Probes Designed to Gene DMG400016227
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26092_PI426222305 | JHI_St_60k_v1 | DMT400041820 | CGAAGATGTAAATGTGATTTCTGTGTTCATCAGATTATATGTACTCCGCGGAATCATATA |
| CUST_26121_PI426222305 | JHI_St_60k_v1 | DMT400041822 | CGAAGATGTAAATGTGATTTCTGTGTTCATCAGATTATATGTACTCCGCGGAATCATATA |
Microarray Signals from CUST_26092_PI426222305
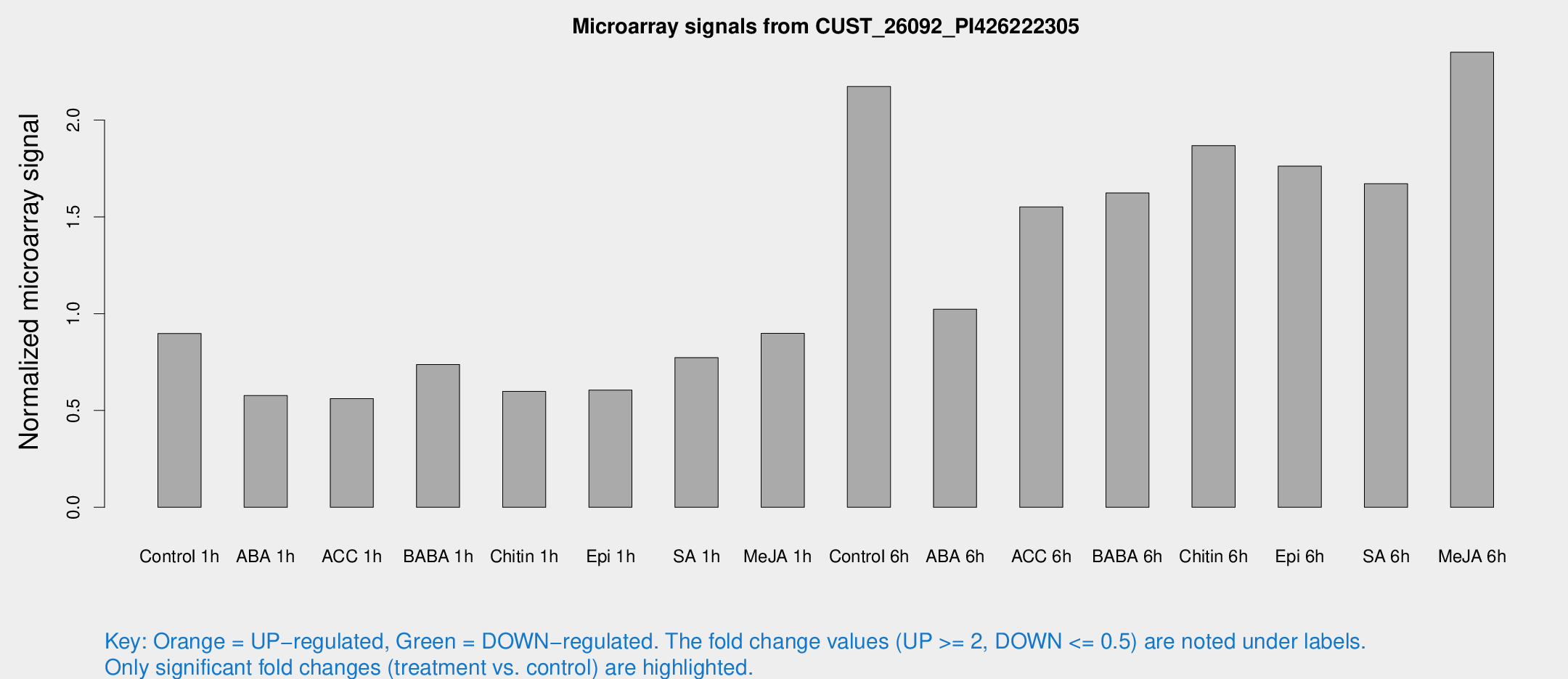
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2562.69 | 261.566 | 0.897508 | 0.0518302 |
| ABA 1h | 1458.56 | 147.606 | 0.577085 | 0.0333413 |
| ACC 1h | 1856.2 | 568.751 | 0.561653 | 0.188477 |
| BABA 1h | 2076.95 | 351.082 | 0.737419 | 0.061207 |
| Chitin 1h | 1526.68 | 88.4562 | 0.598794 | 0.0345941 |
| Epi 1h | 1506.08 | 192.766 | 0.604897 | 0.0776704 |
| SA 1h | 2304.32 | 380.51 | 0.772786 | 0.117156 |
| Me-JA 1h | 2097.22 | 221.495 | 0.898833 | 0.0519132 |
| Control 6h | 6383.39 | 1123.5 | 2.17264 | 0.22009 |
| ABA 6h | 3108.86 | 333.998 | 1.02302 | 0.0645197 |
| ACC 6h | 5068.92 | 450.825 | 1.55089 | 0.106929 |
| BABA 6h | 5159.86 | 348.706 | 1.62293 | 0.0937069 |
| Chitin 6h | 5628.19 | 325.347 | 1.86778 | 0.107843 |
| Epi 6h | 5645.23 | 409.837 | 1.76172 | 0.204165 |
| SA 6h | 4920.44 | 1007.88 | 1.67113 | 0.198241 |
| Me-JA 6h | 6888.3 | 1425.42 | 2.35006 | 0.289469 |
Source Transcript PGSC0003DMT400041820 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G08980.1 | +2 | 2e-43 | 159 | 105/229 (46%) | F-BOX WITH WD-40 2 | chr4:5758993-5760108 FORWARD LENGTH=317 |
