Probe CUST_26030_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26030_PI426222305 | JHI_St_60k_v1 | DMT400052577 | CGTAGTTTGGTTAAACCCACTGAGAATAGACATCACAATTTTTATATCTAAAGACCACAC |
All Microarray Probes Designed to Gene DMG400020401
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26030_PI426222305 | JHI_St_60k_v1 | DMT400052577 | CGTAGTTTGGTTAAACCCACTGAGAATAGACATCACAATTTTTATATCTAAAGACCACAC |
Microarray Signals from CUST_26030_PI426222305
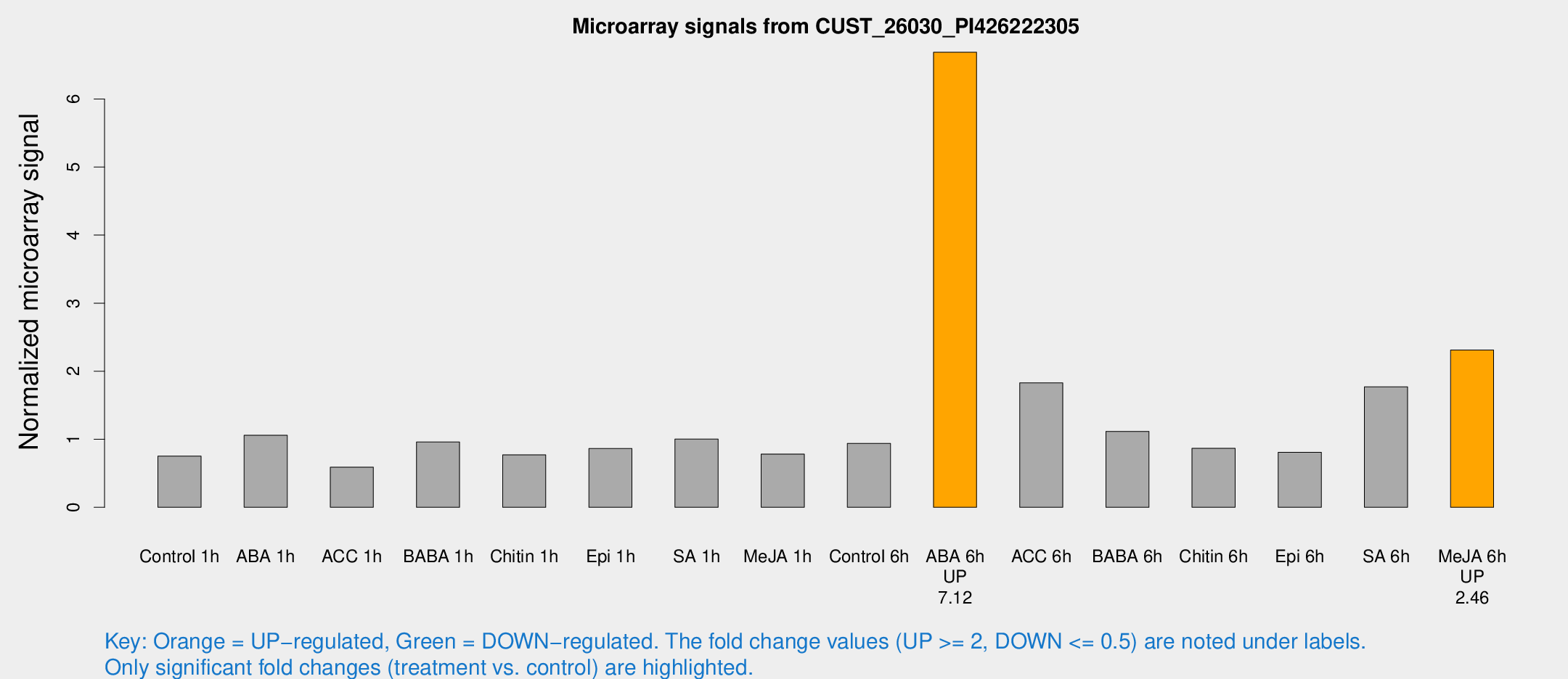
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 47.7519 | 4.32608 | 0.753444 | 0.0811375 |
| ABA 1h | 64.8176 | 20.6405 | 1.05815 | 0.334583 |
| ACC 1h | 43.7965 | 14.767 | 0.589061 | 0.203883 |
| BABA 1h | 60.711 | 11.0108 | 0.960144 | 0.0953731 |
| Chitin 1h | 47.3598 | 14.1449 | 0.770122 | 0.164233 |
| Epi 1h | 48.2821 | 7.71154 | 0.863311 | 0.139578 |
| SA 1h | 66.4571 | 9.6232 | 1.00208 | 0.0956861 |
| Me-JA 1h | 41.8488 | 8.53769 | 0.781993 | 0.0927141 |
| Control 6h | 64.2097 | 17.3969 | 0.93927 | 0.207346 |
| ABA 6h | 470.043 | 92.2321 | 6.6876 | 1.24724 |
| ACC 6h | 134.538 | 16.5962 | 1.82805 | 0.320215 |
| BABA 6h | 81.7697 | 15.5922 | 1.11557 | 0.174816 |
| Chitin 6h | 59.802 | 9.37652 | 0.865769 | 0.126954 |
| Epi 6h | 58.7011 | 8.90247 | 0.807608 | 0.0724573 |
| SA 6h | 111.251 | 9.63188 | 1.76924 | 0.11784 |
| Me-JA 6h | 146.507 | 13.1808 | 2.31209 | 0.353641 |
Source Transcript PGSC0003DMT400052577 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G20700.1 | +3 | 8e-17 | 79 | 79/241 (33%) | Protein of unknown function (DUF581) | chr5:7006178-7007003 REVERSE LENGTH=248 |
