Probe CUST_25942_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25942_PI426222305 | JHI_St_60k_v1 | DMT400051882 | GAAAGAAACATATGGCAAATATTTCTACTTGCTCCCTTCTCGTTGCTGCTGTTTTTCTAG |
All Microarray Probes Designed to Gene DMG400020133
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25942_PI426222305 | JHI_St_60k_v1 | DMT400051882 | GAAAGAAACATATGGCAAATATTTCTACTTGCTCCCTTCTCGTTGCTGCTGTTTTTCTAG |
Microarray Signals from CUST_25942_PI426222305
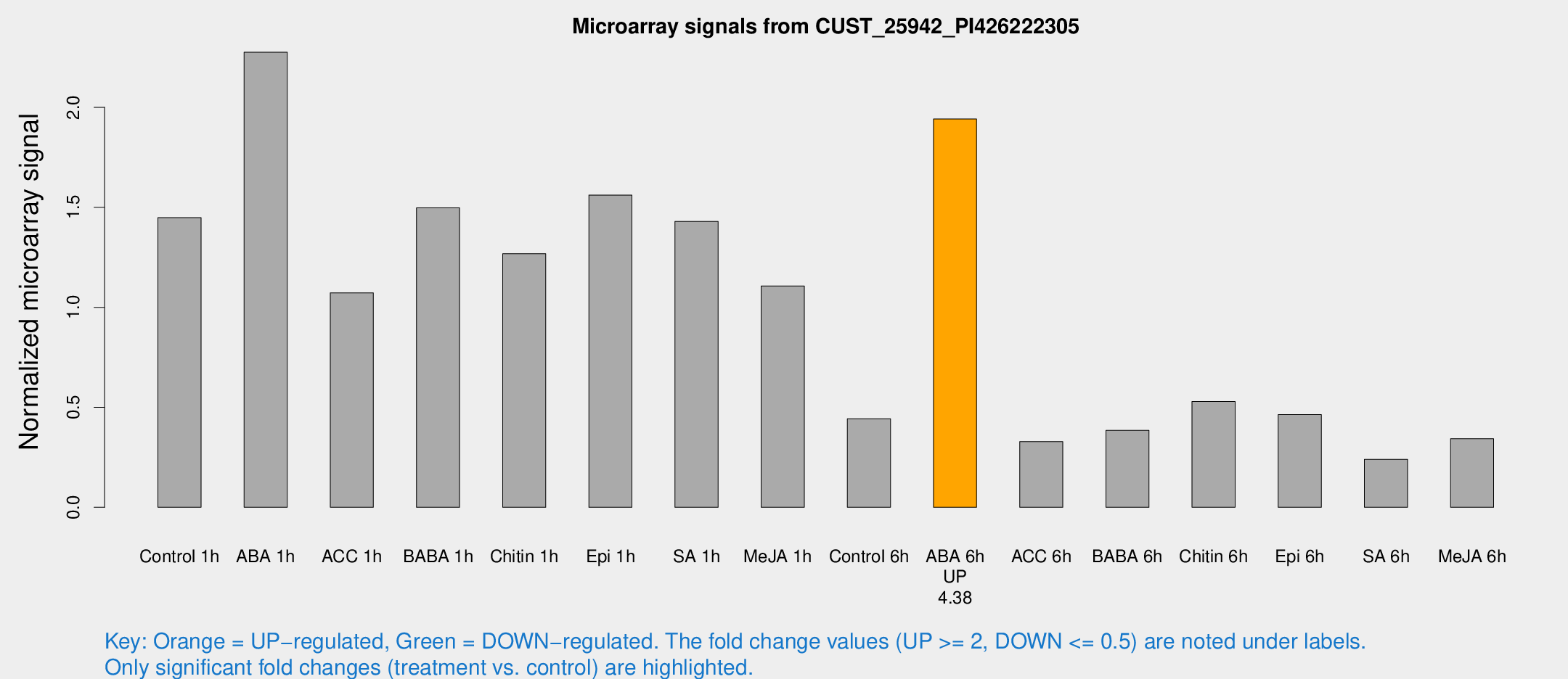
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 135.682 | 9.40205 | 1.44813 | 0.0890765 |
| ABA 1h | 191.015 | 26.059 | 2.27539 | 0.311692 |
| ACC 1h | 128.121 | 46.2624 | 1.07249 | 0.566366 |
| BABA 1h | 136.143 | 14.8544 | 1.4976 | 0.325486 |
| Chitin 1h | 106.472 | 6.78783 | 1.26799 | 0.0872584 |
| Epi 1h | 127.225 | 12.4907 | 1.56093 | 0.150228 |
| SA 1h | 137.124 | 8.43571 | 1.42894 | 0.0939497 |
| Me-JA 1h | 86.0662 | 12.9009 | 1.10669 | 0.244469 |
| Control 6h | 48.1293 | 15.7705 | 0.442877 | 0.159478 |
| ABA 6h | 203.357 | 49.9098 | 1.94139 | 0.371984 |
| ACC 6h | 36.0302 | 6.00923 | 0.328313 | 0.0370697 |
| BABA 6h | 42.1023 | 9.59793 | 0.384338 | 0.087997 |
| Chitin 6h | 52.5849 | 4.45656 | 0.529022 | 0.0449605 |
| Epi 6h | 50.99 | 9.98401 | 0.463305 | 0.144592 |
| SA 6h | 24.6292 | 8.47516 | 0.240004 | 0.0690222 |
| Me-JA 6h | 33.3812 | 7.53892 | 0.342695 | 0.0620656 |
Source Transcript PGSC0003DMT400051882 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
