Probe CUST_2572_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2572_PI426222305 | JHI_St_60k_v1 | DMT400072451 | GATTTTGGAAATCTAAGATTGGATATTAAGATAGTTCCTTCTATGCTAGCTAGAGCTAGG |
All Microarray Probes Designed to Gene DMG400028186
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2550_PI426222305 | JHI_St_60k_v1 | DMT400072450 | TCCAACAAAAGGGTGTTCTTGAAGTTCCTTGTAGCGCCGCGGAATTGGAAAAGATTTTGG |
| CUST_2572_PI426222305 | JHI_St_60k_v1 | DMT400072451 | GATTTTGGAAATCTAAGATTGGATATTAAGATAGTTCCTTCTATGCTAGCTAGAGCTAGG |
Microarray Signals from CUST_2572_PI426222305
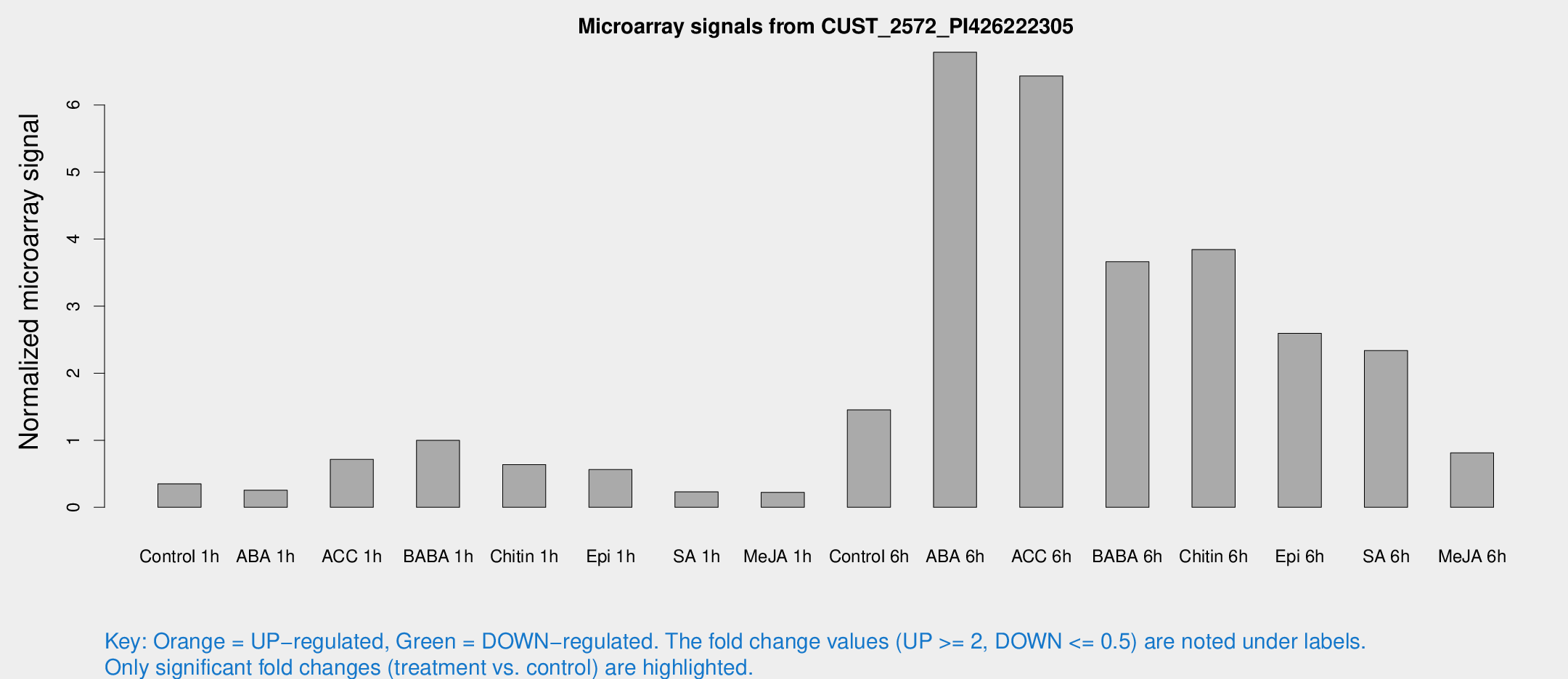
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 16.1576 | 3.80059 | 0.350429 | 0.0893676 |
| ABA 1h | 12.2008 | 5.32851 | 0.256307 | 0.118683 |
| ACC 1h | 43.2432 | 17.9315 | 0.713738 | 0.439848 |
| BABA 1h | 49.6542 | 19.021 | 0.998732 | 0.316239 |
| Chitin 1h | 25.905 | 3.92584 | 0.637609 | 0.0975172 |
| Epi 1h | 24.7454 | 7.82099 | 0.56281 | 0.223792 |
| SA 1h | 10.6074 | 3.6631 | 0.229597 | 0.0796879 |
| Me-JA 1h | 9.32333 | 3.8215 | 0.223582 | 0.108605 |
| Control 6h | 90.3693 | 38.9137 | 1.45284 | 1.02571 |
| ABA 6h | 410.24 | 206.096 | 6.78641 | 4.35931 |
| ACC 6h | 361.031 | 109.557 | 6.433 | 1.07179 |
| BABA 6h | 200.399 | 61.0752 | 3.66171 | 0.957646 |
| Chitin 6h | 193.269 | 45.258 | 3.84363 | 0.891903 |
| Epi 6h | 136.247 | 24.2406 | 2.59495 | 0.731821 |
| SA 6h | 105.452 | 12.9993 | 2.33798 | 0.208728 |
| Me-JA 6h | 49.1817 | 22.6665 | 0.81265 | 0.519786 |
Source Transcript PGSC0003DMT400072451 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G09870.1 | +3 | 1e-18 | 80 | 46/109 (42%) | SAUR-like auxin-responsive protein family | chr3:3027555-3027896 REVERSE LENGTH=113 |
