Probe CUST_25691_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25691_PI426222305 | JHI_St_60k_v1 | DMT400037294 | AGACATTGTATCTTTCTACCAGTATAGTGATGCTATCAGTTTGTAATTTGACGTTTGTCG |
All Microarray Probes Designed to Gene DMG400014391
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25691_PI426222305 | JHI_St_60k_v1 | DMT400037294 | AGACATTGTATCTTTCTACCAGTATAGTGATGCTATCAGTTTGTAATTTGACGTTTGTCG |
Microarray Signals from CUST_25691_PI426222305
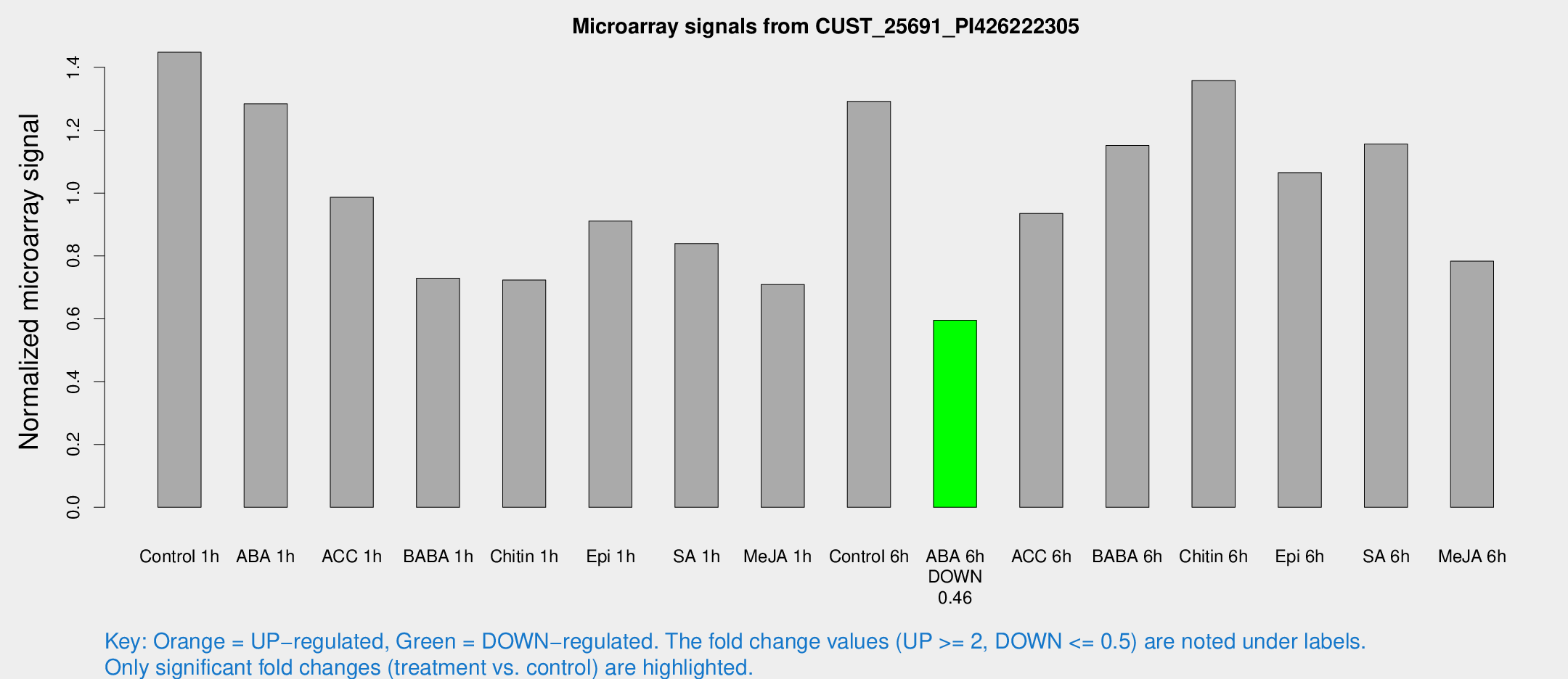
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2714.21 | 451.872 | 1.44815 | 0.140148 |
| ABA 1h | 2089.02 | 182.248 | 1.28444 | 0.132078 |
| ACC 1h | 1979.31 | 468.668 | 0.986685 | 0.201961 |
| BABA 1h | 1392.05 | 353.781 | 0.729041 | 0.149025 |
| Chitin 1h | 1203.48 | 137.535 | 0.723221 | 0.0883885 |
| Epi 1h | 1463.03 | 179.064 | 0.910901 | 0.109217 |
| SA 1h | 1611.14 | 256.69 | 0.839215 | 0.0928029 |
| Me-JA 1h | 1077.29 | 154.882 | 0.709013 | 0.0431757 |
| Control 6h | 2477.48 | 525.542 | 1.29192 | 0.199379 |
| ABA 6h | 1158.66 | 67.917 | 0.595114 | 0.0344081 |
| ACC 6h | 2095.71 | 485.597 | 0.935131 | 0.199881 |
| BABA 6h | 2415.78 | 408.474 | 1.15145 | 0.188745 |
| Chitin 6h | 2707.7 | 448.464 | 1.35813 | 0.175094 |
| Epi 6h | 2196.88 | 127.18 | 1.06519 | 0.0629029 |
| SA 6h | 2216.82 | 486.796 | 1.15624 | 0.162369 |
| Me-JA 6h | 1517.9 | 375.672 | 0.783098 | 0.137327 |
Source Transcript PGSC0003DMT400037294 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G42550.1 | +3 | 0.0 | 787 | 470/907 (52%) | plastid movement impaired1 | chr1:15977131-15979734 FORWARD LENGTH=843 |
